Generation and release of nitrotyrosine O-sulfate by HepG2 human hepatoma cells upon SIN-1 stimulation: identification of SULT1A3 as the enzyme responsible
- PMID: 17002600
- PMCID: PMC1820819
- DOI: 10.1042/BJ20060536
Generation and release of nitrotyrosine O-sulfate by HepG2 human hepatoma cells upon SIN-1 stimulation: identification of SULT1A3 as the enzyme responsible
Abstract
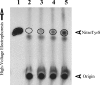
In addition to serving as a biomarker of oxidative/nitrative stress, elevated levels of nitrotyrosine have been shown to cause DNA damage or trigger apoptosis. Whether the body is equipped with mechanisms for protecting against the potentially harmful nitrotyrosine remains unknown. The present study was designed to investigate the possibility that sulfation serves as a pathway for the metabolism/regulation of nitrotyrosine. Using metabolic labelling, nitrotyrosine O-[35S]sulfate was found to be produced and released into the medium of HepG2 human hepatoma cells labelled with [35S]sulfate in the presence of nitrotyrosine. To identify the enzyme(s) responsible for nitrotyrosine sulfation, a systematic study of all eleven known human cytosolic SULTs (sulfotransferases) was performed. Of the 11 enzymes tested, only SULT1A3 displayed sulfating activity toward nitrotyrosine. The pH-dependence and kinetic constants of SULT1A3 with nitrotyrosine or dopamine as substrate were determined. To examine whether the sulfation of nitrotyrosine occurs in the context of cellular physiology, HepG2 cells labelled with [35S]sulfate were treated with SIN-1 (morpholinosydnonimine), a peroxynitrite generator. Increments of nitrotyrosine O-[35S]sulfate were detected in the medium of HepG2 cells treated with higher concentrations of SIN-1. To gain insight into the physiological relevance of nitrotyrosine sulfation, a time-course study was performed using [3H]tyrosine-labelled HepG2 cells treated with SIN-1. The findings confirm that the bulk of free [3H]nitrotyrosine inside the cells was present in the unconjugated form. The proportion of sulfated [3H]nitrotyrosine increased dramatically in the medium over time, implying that sulfation may play a significant role in the metabolism of free nitrotyrosine.
Figures

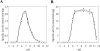


Similar articles
-
Sulfation of nitrotyrosine: biochemistry and functional implications.IUBMB Life. 2007 Oct;59(10):622-7. doi: 10.1080/15216540701589320. IUBMB Life. 2007. PMID: 17891604 Review.
-
Sulfation of chlorotyrosine and nitrotyrosine by human lung endothelial and epithelial cells: role of the human SULT1A3.Toxicol Appl Pharmacol. 2011 Mar 1;251(2):104-9. doi: 10.1016/j.taap.2010.12.006. Epub 2010 Dec 17. Toxicol Appl Pharmacol. 2011. PMID: 21168432
-
Hydroxylated serotonin and dopamine as substrates and inhibitors for human cytosolic SULT1A3.J Neurochem. 2007 Dec;103(6):2679-89. doi: 10.1111/j.1471-4159.2007.04948.x. J Neurochem. 2007. PMID: 17908235
-
De novo sulfation of L-tyrosine in HepG2 human hepatoma cells and its possible functional implication.Eur J Biochem. 1994 Dec 1;226(2):293-301. doi: 10.1111/j.1432-1033.1994.tb20053.x. Eur J Biochem. 1994. PMID: 8001547
-
Sulfation of drugs and hormones in mid-gestation human fetus.Early Hum Dev. 2005 Jul;81(7):573-81. doi: 10.1016/j.earlhumdev.2004.10.021. Epub 2004 Dec 8. Early Hum Dev. 2005. PMID: 16009282 Review.
Cited by
-
Co-inhibition of HDAC and MLL-menin interaction targets MLL-rearranged acute myeloid leukemia cells via disruption of DNA damage checkpoint and DNA repair.Clin Epigenetics. 2019 Oct 7;11(1):137. doi: 10.1186/s13148-019-0723-0. Clin Epigenetics. 2019. PMID: 31590682 Free PMC article.
-
SULT genetic polymorphisms: physiological, pharmacological and clinical implications.Expert Opin Drug Metab Toxicol. 2021 Jul;17(7):767-784. doi: 10.1080/17425255.2021.1940952. Epub 2021 Jun 30. Expert Opin Drug Metab Toxicol. 2021. PMID: 34107842 Free PMC article. Review.
-
Comparative assessment of phototherapy protocols for reduction of oxidative stress in partially transected spinal cord slices undergoing secondary degeneration.BMC Neurosci. 2016 May 18;17(1):21. doi: 10.1186/s12868-016-0259-6. BMC Neurosci. 2016. PMID: 27194427 Free PMC article.
References
-
- Ischiropoulos H. Biological tyrosine nitration: a pathophysiological function of nitric oxide and reactive oxygen species. Arch. Biochem. Biophys. 1998;356:1–11. - PubMed
-
- Beckmann J. S., Ye Y. Z., Anderson P. G., Chen J., Accavitti M. A., Tarpey M. M., White C. R. Extensive nitration of protein tyrosines in human atherosclerosis detected by immunohistochemistry. Biol. Chem. Hoppe Seyler. 1994;375:81–88. - PubMed
-
- Cromheeke K. M., Kockx M. M., De Meyer G. R., Bosmans J. M., Bult H., Beelaerts W. J., Vrints C. J., Herman A. G. Inducible nitric oxide synthase colocalizes with signs of lipid oxidation/peroxidation in human atherosclerotic plaques. Cardiovasc. Res. 1999;43:744–754. - PubMed
-
- Forster C., Clark H. B., Ross M. E., Iadecola C. Inducible nitric oxide synthase expression in human cerebral infarcts. Acta Neuropathol. 1999;97:215–220. - PubMed
-
- Andreadis A. A., Hazen S. L., Comhair S. A., Erzurum S. C. Oxidative and nitrosative events in asthma. Free Radical Biol. Med. 2003;35:213–225. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

