HIV-1 designed to use different tRNAGln isoacceptors prefers to select tRNAThr for replication
- PMID: 17002807
- PMCID: PMC1592299
- DOI: 10.1186/1743-422X-3-80
HIV-1 designed to use different tRNAGln isoacceptors prefers to select tRNAThr for replication
Abstract
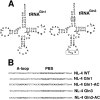
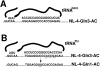
Background: Previous studies have shown that infection with human immunodeficiency virus type 1 (HIV-1) causes acceleration of the synthesis of glutamine tRNA (tRNAGln) in infected cells. To investigate whether this might influence HIV-1 to utilize tRNAGln as a primer for initiation of reverse transcription, we have constructed HIV-1 proviral genomes in which the PBS and the A-loop region upstream of the PBS have been made complementary to either the anticodon region of tRNAGln,1 or tRNAGln,3 and 3' terminal 18 nucleotides of each isoacceptor of tRNAGln.
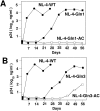
Results: Viruses in which the PBS was altered to be complementary to tRNAGln,1 or tRNAGln,3 with or without the A-loop all exhibited a lower infectivity than the wild type virus. Viruses with only the PBS complementary to tRNAGln,1 or tRNAGln,3 reverted to wild type following culture in SupT1 cells. Surprisingly, viruses in which the PBS and A-loop were complementary to tRNAGln,1 did not grow in SupT1 cells, while viruses in which the PBS and A-loop were made complementary to tRNAGln,3 grew slowly in SupT1 cells. Analysis of the PBS of this virus revealed that it had reverted to select tRNAThr as the primer, which shares complementarity in 15 of 18 nucleotides with the PBS complementary to tRNAGln,3.
Conclusion: The results of these studies support the concept that the HIV-1 has preferred tRNAs that can be selected as primers for replication.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

