Legume genome evolution viewed through the Medicago truncatula and Lotus japonicus genomes
- PMID: 17003129
- PMCID: PMC1578499
- DOI: 10.1073/pnas.0603228103
Legume genome evolution viewed through the Medicago truncatula and Lotus japonicus genomes
Erratum in
- Proc Natl Acad Sci U S A. 2006 Nov 21;103(47):18026. Scheix, Thomas [corrected to Schiex, Thomas]
Abstract
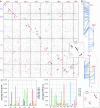
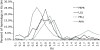
Genome sequencing of the model legumes, Medicago truncatula and Lotus japonicus, provides an opportunity for large-scale sequence-based comparison of two genomes in the same plant family. Here we report synteny comparisons between these species, including details about chromosome relationships, large-scale synteny blocks, microsynteny within blocks, and genome regions lacking clear correspondence. The Lotus and Medicago genomes share a minimum of 10 large-scale synteny blocks, each with substantial collinearity and frequently extending the length of whole chromosome arms. The proportion of genes syntenic and collinear within each synteny block is relatively homogeneous. Medicago-Lotus comparisons also indicate similar and largely homogeneous gene densities, although gene-containing regions in Mt occupy 20-30% more space than Lj counterparts, primarily because of larger numbers of Mt retrotransposons. Because the interpretation of genome comparisons is complicated by large-scale genome duplications, we describe synteny, synonymous substitutions and phylogenetic analyses to identify and date a probable whole-genome duplication event. There is no direct evidence for any recent large-scale genome duplication in either Medicago or Lotus but instead a duplication predating speciation. Phylogenetic comparisons place this duplication within the Rosid I clade, clearly after the split between legumes and Salicaceae (poplar).
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

