Repression of HMGA2 gene expression by human cytomegalovirus involves the IE2 86-kilodalton protein and is necessary for efficient viral replication and inhibition of cyclin A transcription
- PMID: 17005673
- PMCID: PMC1617307
- DOI: 10.1128/JVI.01300-06
Repression of HMGA2 gene expression by human cytomegalovirus involves the IE2 86-kilodalton protein and is necessary for efficient viral replication and inhibition of cyclin A transcription
Abstract
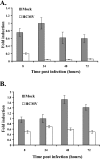
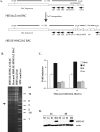
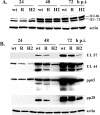
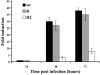
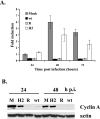
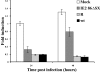
Human cytomegalovirus (HCMV) infection results in dysregulation of several cell cycle genes, including inhibition of cyclin A transcription. In this work, we examine the effect of the HCMV infection on expression of the high-mobility group A2 (HMGA2) gene, which encodes an architectural transcription factor that is involved in cyclin A promoter activation. We find that expression of HMGA2 RNA is repressed in infected cells. To determine whether repression of HMGA2 is directly related to the inhibition of cyclin A expression and impacts on the progression of the infection, we constructed an HCMV recombinant that expressed HMGA2. In cells infected with the recombinant virus, cyclin A mRNA and protein are induced, and there is a significant delay in viral early gene expression and DNA replication. To determine the mechanism of HMGA2 repression, we used recombinant viruses that expressed either no IE1 72-kDa protein (CR208) or greatly reduced levels of IE2 86-kDa (IE2 86) protein (IE2 86DeltaSX-EGFP). At a high multiplicity of infection, the IE1 deletion mutant is comparable to the wild type with respect to inhibition of HMGA2. In contrast, the IE2 86DeltaSX-EGFP mutant does not significantly repress HMGA2 expression, suggesting that IE2 86 is involved in the regulation of this gene. Cyclin A expression is also induced in cells infected with this mutant virus. Since HMGA2 is important for cell proliferation and differentiation, particularly during embryogenesis, it is possible that the repression of HMGA2 expression during fetal development could contribute to the specific birth defects in HCMV-infected neonates.
Figures
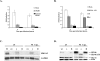
References
-
- Anand, A., and K. Chada. 2000. In vivo modulation of Hmgic reduces obesity. Nat. Genet. 24:377-380. - PubMed
-
- Arlotta, P., A. K. Tai, G. Manfioletti, C. Clifford, G. Jay, and S. J. Ono. 2000. Transgenic mice expressing a truncated form of the high mobility group I-C protein develop adiposity and an abnormally high prevalence of lipomas. J. Biol. Chem. 275:14394-14400. - PubMed
-
- Baldassarre, G., M. Fedele, S. Battista, A. Vecchione, A. J. Klein-Szanto, M. Santoro, T. A. Waldmann, N. Azimi, C. M. Croce, and A. Fusco. 2001. Onset of natural killer cell lymphomas in transgenic mice carrying a truncated HMGI-C gene by the chronic stimulation of the IL-2 and IL-15 pathway. Proc. Natl. Acad. Sci. USA 98:7970-7975. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

