Autorepression of the human cytomegalovirus major immediate-early promoter/enhancer at late times of infection is mediated by the recruitment of chromatin remodeling enzymes by IE86
- PMID: 17005678
- PMCID: PMC1617317
- DOI: 10.1128/JVI.01297-06
Autorepression of the human cytomegalovirus major immediate-early promoter/enhancer at late times of infection is mediated by the recruitment of chromatin remodeling enzymes by IE86
Abstract
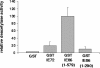
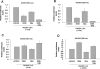
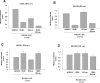
The human cytomegalovirus major immediate-early protein IE86 is pivotal for coordinated regulation of viral gene expression throughout infection. A relatively promiscuous transactivator of viral early and late gene transcription, IE86 also acts during infection to negatively regulate its own promoter via direct binding to a 14-bp palindromic IE86-binding site, the cis repression sequence (crs), located between the major immediate-early promoter (MIEP) TATA box and the start of transcription. Although such autoregulation does not involve changes in the binding of basal transcription factors to the MIEP in vitro, it does appear to involve selective inhibition of RNA polymerase II recruitment. However, how this occurs is unclear. We show that autorepression by IE86 at late times of infection correlates with changes in chromatin structure around the MIEP during the course of infection and that this is likely to result from physical and functional interactions between IE86 and chromatin remodeling enzymes normally associated with transcriptional repression of cellular promoters. Firstly, we show that IE86-mediated autorepression is inhibited by histone deacetylase inhibitors. We also show that IE86 interacts, in vitro and in vivo, with the histone deacetylase HDAC1 and histone methyltransferases G9a and Suvar(3-9)H1 and that coexpression of these chromatin remodeling enzymes with IE86 increases autorepression of the MIEP. Finally, we show that mutation of the crs in the context of the virus abrogates the transcriptionally repressive chromatin phenotype normally found around the MIEP at late times of infection, suggesting that negative autoregulation by IE86 results, at least in part, from IE86-mediated changes in chromatin structure of the viral MIEP.
Figures






Similar articles
-
Site-specific inhibition of RNA polymerase II preinitiation complex assembly by human cytomegalovirus IE86 protein.J Virol. 1993 Dec;67(12):7547-55. doi: 10.1128/JVI.67.12.7547-7555.1993. J Virol. 1993. PMID: 8230474 Free PMC article.
-
The human cytomegalovirus 86-kilodalton major immediate-early protein interacts physically and functionally with histone acetyltransferase P/CAF.J Virol. 2000 Aug;74(16):7230-7. doi: 10.1128/jvi.74.16.7230-7237.2000. J Virol. 2000. PMID: 10906177 Free PMC article.
-
Ets-2 repressor factor recruits histone deacetylase to silence human cytomegalovirus immediate-early gene expression in non-permissive cells.J Gen Virol. 2005 Mar;86(Pt 3):535-544. doi: 10.1099/vir.0.80352-0. J Gen Virol. 2005. PMID: 15722512
-
Chromatin-mediated regulation of cytomegalovirus gene expression.Virus Res. 2011 May;157(2):134-43. doi: 10.1016/j.virusres.2010.09.019. Epub 2010 Sep 25. Virus Res. 2011. PMID: 20875471 Free PMC article. Review.
-
Chromatin structure regulates human cytomegalovirus gene expression during latency, reactivation and lytic infection.Biochim Biophys Acta. 2010 Mar-Apr;1799(3-4):286-95. doi: 10.1016/j.bbagrm.2009.08.001. Epub 2009 Aug 12. Biochim Biophys Acta. 2010. PMID: 19682613 Review.
Cited by
-
Control of Immediate Early Gene Expression for Human Cytomegalovirus Reactivation.Front Cell Infect Microbiol. 2020 Sep 17;10:476. doi: 10.3389/fcimb.2020.00476. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 33072616 Free PMC article. Review.
-
Quantitative proteomic discovery of dynamic epigenome changes that control human cytomegalovirus (HCMV) infection.Mol Cell Proteomics. 2014 Sep;13(9):2399-410. doi: 10.1074/mcp.M114.039792. Epub 2014 Jul 1. Mol Cell Proteomics. 2014. PMID: 24987098 Free PMC article.
-
Regulation of human cytomegalovirus transcription in latency: beyond the major immediate-early promoter.Viruses. 2013 Jun 3;5(6):1395-413. doi: 10.3390/v5061395. Viruses. 2013. PMID: 23736881 Free PMC article. Review.
-
Human cytomegalovirus pUL97 regulates the viral major immediate early promoter by phosphorylation-mediated disruption of histone deacetylase 1 binding.J Virol. 2013 Jul;87(13):7393-408. doi: 10.1128/JVI.02825-12. Epub 2013 Apr 24. J Virol. 2013. PMID: 23616659 Free PMC article.
-
Chromatin control of human cytomegalovirus infection.mBio. 2023 Aug 31;14(4):e0032623. doi: 10.1128/mbio.00326-23. Epub 2023 Jul 13. mBio. 2023. PMID: 37439556 Free PMC article. Review.
References
-
- Asmar, J., L. Wiebusch, M. Truss, and C. Hagemeier. 2004. The putative zinc finger of the human cytomegalovirus IE2 86-kilodalton protein is dispensable for DNA binding and autorepression, thereby demarcating a concise core domain in the C terminus of the protein. J. Virol. 78:11853-11864. - PMC - PubMed
-
- Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire, and T. Kouzarides. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120-124. - PubMed
-
- Berger, S. L. 2002. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12:142-148. - PubMed
-
- Bottardi, S., A. Aumont, F. Grosveld, and E. Milot. 2003. Developmental stage-specific epigenetic control of human beta-globin gene expression is potentiated in hematopoietic progenitor cells prior to their transcriptional activation. Blood 102:3989-3997. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

