RNA silencing suppression by a second copy of the P1 serine protease of Cucumber vein yellowing ipomovirus, a member of the family Potyviridae that lacks the cysteine protease HCPro
- PMID: 17005683
- PMCID: PMC1617295
- DOI: 10.1128/JVI.00985-06
RNA silencing suppression by a second copy of the P1 serine protease of Cucumber vein yellowing ipomovirus, a member of the family Potyviridae that lacks the cysteine protease HCPro
Abstract
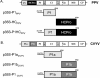
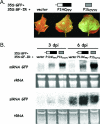
The P1 protein of viruses of the family Potyviridae is a serine proteinase, which is highly variable in length and sequence, and its role in the virus infection cycle is not clear. One of the proposed activities of P1 is to assist HCPro, the product that viruses of the genus Potyvirus use to counteract antiviral defense mediated by RNA silencing. Indeed, an HCPro-coding region is present in all the genomes of members of the genera Potyvirus, Rymovirus, and Tritimovirus that have been sequenced. However, it was recently reported that a sequence coding for HCPro is lacking in the genome of Cucumber vein yellowing virus (CVYV), a member of the genus Ipomovirus, the fourth monopartite genus of the family. In this study, we provide further evidence that P1 enhances the activity of HCPro in members of the genus Potyvirus and show that it is duplicated in the ipomovirus CVYV. The two CVYV P1 copies are arranged in tandem, and the second copy (P1b) has RNA silencing suppression activity. CVYV P1b suppressed RNA silencing induced either by sense green fluorescent protein (GFP) mRNA or by a GFP inverted repeat RNA, indicating that CVYV P1b acts downstream of the formation of double-stranded RNA. CVYV P1b also suppressed local silencing in agroinfiltrated patches of transgenic Nicotiana benthamiana line 16c and delayed its propagation to the neighboring cells. However, neither the short-distance nor long-distance systemic spread of silencing of the GFP transgene was completely blocked by CVYV P1b. CVYV P1b and P1-HCPro from the potyvirus Plum pox virus showed very similar behaviors in all the assays carried out, suggesting that evolution has found a way to counteract RNA silencing by similar mechanisms using very different proteins in viruses of the same family.
Figures




References
-
- Adams, M. J., J. F. Antoniw, and C. M. Fauquet. 2005. Molecular criteria for genus and species discrimination within the family Potyviridae. Arch. Virol. 150:459-479. - PubMed
-
- Baulcombe, D. 2004. RNA silencing in plants. Nature 431:356-363. - PubMed
-
- Bendahmane, A., G. Farnham, P. Moffett, and D. C. Baulcombe. 2002. Constitutive gain-of-function mutants in a nucleotide binding site-leucine rich repeat protein encoded at the Rx locus of potato. Plant J. 32:195-204. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

