Packaging of brome mosaic virus subgenomic RNA is functionally coupled to replication-dependent transcription and translation of coat protein
- PMID: 17005687
- PMCID: PMC1617292
- DOI: 10.1128/JVI.01186-06
Packaging of brome mosaic virus subgenomic RNA is functionally coupled to replication-dependent transcription and translation of coat protein
Abstract
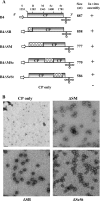
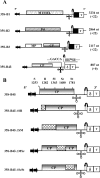
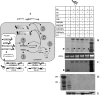
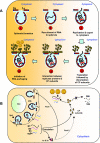
In Brome mosaic virus (BMV), genomic RNA1 (gB1) and RNA2 (gB2), encoding the replication factors, are packaged into two separate virions, whereas genomic RNA3 (gB3) and its subgenomic coat protein (CP) mRNA (sgB4) are copackaged into a third virion. In vitro assembly assays performed between a series of deletion variants of sgB4 and wild-type (wt) CP subunits demonstrated that packaging of sgB4 is independent of sequences encoding the CP open reading frame. To confirm these observations in vivo and to unravel the mechanism of sgB4 copackaging, an Agrobacterium-mediated transient in vivo expression system (P. Annamalai and A. L. N. Rao, Virology 338:96-111, 2005) that effectively uncouples replication from packaging was used. Cultures of agrotransformants, engineered to express sgB4 and CP subunits either transiently (sgB4(Trans) and CP(Trans)) or in replication-dependent transcription and translation when complemented with gB1 and gB2 (sgB4(Rep) and CP(Rep)), were mixed in all four pair-wise combinations and infiltrated to Nicotiana benthamiana leaves to systematically evaluate requirements regulating sgB4 packaging. The data revealed that (i) in the absence of replication, packaging was nonspecific, since transiently expressed CP subunits efficiently packaged ubiquitous cellular RNA as well as transiently expressed sgB4 and its deletion variants; (ii) induction of viral replication increased specificity of RNA packaging; and most importantly, (iii) efficient packaging of sgB4, reminiscent of the wt scenario, is functionally coupled not only to its transcription via replication but also to translation of CP from replication-derived mRNA, a mechanism that appears to be conserved among positive-strand RNA viruses of plants (this study), animals (flock house virus), and humans (poliovirus).
Figures





References
-
- Annamalai, P., and A. L. Rao. 2006. Delivery and expression of functional viral RNA genomes in planta by agroinfiltration, p. 16B.2.1-2.15. In T. Downey (ed.), Current protocols in microbiology, vol. 1. John Wiley & Sons, Inc., Hoboken, N.J. - PubMed
-
- Annamalai, P., and A. L. Rao. 2005. Replication-independent expression of genome components and capsid protein of brome mosaic virus in planta: a functional role for viral replicase in RNA packaging. Virology 338:96-111. - PubMed
-
- Basnayake, V. R., T. L. Sit, and S. A. Lommel. 2006. The genomic RNA packaging scheme of Red clover necrotic mosaic virus. Virology 345:532-539. - PubMed
-
- Berkowitz, R., J. Fisher, and S. P. Goff. 1996. RNA packaging. Curr. Top. Microbiol. Immunol. 214:177-218. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

