Identification of a novel oligodendrocyte cell adhesion protein using gene expression profiling
- PMID: 17005852
- PMCID: PMC1613258
- DOI: 10.1523/JNEUROSCI.2246-06.2006
Identification of a novel oligodendrocyte cell adhesion protein using gene expression profiling
Abstract
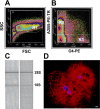
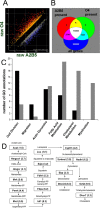
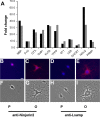
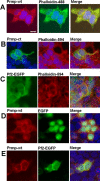
Oligodendrocytes undergo extensive changes as they differentiate from progenitors into myelinating cells. To better understand the molecular mechanisms underlying this transformation, we performed a comparative analysis using gene expression profiling of A2B5+ oligodendrocyte progenitors and O4+ oligodendrocytes. Cells were sort-purified ex vivo from postnatal rat brain using flow cytometry. Using Affymetrix microarrays, 1707 transcripts were identified with a more than twofold increase in expression in O4+ oligodendrocytes. Many genes required for oligodendrocyte differentiation were upregulated in O4+ oligodendrocytes, including numerous genes encoding myelin proteins. Transcriptional changes included genes required for cell adhesion, actin cytoskeleton regulation, and fatty acid and cholesterol biosynthesis. At the O4+ stage, there was an increase in expression of a novel proline-rich transmembrane protein (Prmp). Localized to the plasma membrane, Prmp displays adhesive properties that may be important for linking the extracellular matrix to the actin cytoskeleton. Together, our results highlight the usefulness of this discovery-driven experimental strategy to identify genes relevant to oligodendrocyte differentiation and myelination.
Figures


Similar articles
-
Non-cell autonomous impairment of oligodendrocyte differentiation precedes CNS degeneration in the Zitter rat: implications of macrophage/microglial activation in the pathogenesis.BMC Neurosci. 2008 Apr 5;9:35. doi: 10.1186/1471-2202-9-35. BMC Neurosci. 2008. PMID: 18394170 Free PMC article.
-
Phosphodiesterase I, a novel adhesion molecule and/or cytokine involved in oligodendrocyte function.J Neurosci. 1997 Dec 1;17(23):9095-103. doi: 10.1523/JNEUROSCI.17-23-09095.1997. J Neurosci. 1997. PMID: 9364056 Free PMC article.
-
The tetraspanin protein, CD9, is expressed by progenitor cells committed to oligodendrogenesis and is linked to beta1 integrin, CD81, and Tspan-2.Glia. 2002 Dec;40(3):350-9. doi: 10.1002/glia.10134. Glia. 2002. PMID: 12420314
-
Mechanical plasticity during oligodendrocyte differentiation and myelination.Glia. 2018 Jan;66(1):5-14. doi: 10.1002/glia.23206. Epub 2017 Sep 21. Glia. 2018. PMID: 28940651 Free PMC article. Review.
-
Exploring oligodendrocyte guidance: 'to boldly go where no cell has gone before'.Cell Mol Life Sci. 2005 Mar;62(5):505-10. doi: 10.1007/s00018-004-4485-1. Cell Mol Life Sci. 2005. PMID: 15747057 Free PMC article. Review.
Cited by
-
Flow cytometry protocols for surface and intracellular antigen analyses of neural cell types.J Vis Exp. 2014 Dec 18;(94):52241. doi: 10.3791/52241. J Vis Exp. 2014. PMID: 25549236 Free PMC article.
-
Transcriptome analysis identifies genes with enriched expression in the mouse central extended amygdala.Neuroscience. 2008 Oct 28;156(4):950-65. doi: 10.1016/j.neuroscience.2008.07.070. Epub 2008 Aug 14. Neuroscience. 2008. PMID: 18786617 Free PMC article.
-
The calcium-dependent interaction between S100B and the mitochondrial AAA ATPase ATAD3A and the role of this complex in the cytoplasmic processing of ATAD3A.Mol Cell Biol. 2010 Jun;30(11):2724-36. doi: 10.1128/MCB.01468-09. Epub 2010 Mar 29. Mol Cell Biol. 2010. PMID: 20351179 Free PMC article.
-
Hippocampal CA3 transcriptome signature correlates with initial precipitating injury in refractory mesial temporal lobe epilepsy.PLoS One. 2011;6(10):e26268. doi: 10.1371/journal.pone.0026268. Epub 2011 Oct 14. PLoS One. 2011. PMID: 22022585 Free PMC article.
-
Sox10 cooperates with the mediator subunit 12 during terminal differentiation of myelinating glia.J Neurosci. 2013 Apr 10;33(15):6679-90. doi: 10.1523/JNEUROSCI.5178-12.2013. J Neurosci. 2013. PMID: 23575864 Free PMC article.
References
-
- Abney ER, Williams BP, Raff MC. Tracing the development of oligodendrocytes from precursor cells using monoclonal antibodies, fluorescence-activated cell sorting, and cell culture. Dev Biol. 1983;100:66–171. - PubMed
-
- Armstrong RC. Isolation and characterization of immature oligodendrocyte lineage cells. Methods. 1998;16:282–292. - PubMed
-
- Asou H, Hamada K, Miyazaki T, Sakota T, Hayashi K, Takeda Y, Marret S, Delpech B, Itoh K, Uyemura K. CNS myelinogenesis in vitro: time course and pattern of rat oligodendrocyte development. J Neurosci Res. 1995;40:519–534. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
