Vestibular hair bundles control pH with (Na+, K+)/H+ exchangers NHE6 and NHE9
- PMID: 17005858
- PMCID: PMC6674470
- DOI: 10.1523/JNEUROSCI.2990-06.2006
Vestibular hair bundles control pH with (Na+, K+)/H+ exchangers NHE6 and NHE9
Abstract
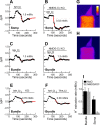
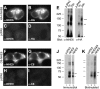
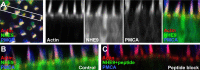
In hair cells of the inner ear, robust Ca2+/H+ exchange mediated by plasma-membrane Ca2+-ATPase would rapidly acidify mechanically sensitive hair bundles without efficient removal of H+. We found that, whereas the basolateral membrane of vestibular hair cells from the frog saccule extrudes H+ via an Na+-dependent mechanism, bundles rapidly remove H+ in the absence of Na+ and HCO3(-), even when the soma is acidified. K+ was fully effective and sufficient for H+ removal; in contrast, Rb+ failed to support pH recovery. Na+/H+-exchanger isoform 1 (NHE1) was present on hair-cell soma membranes and was likely responsible for Na+-dependent H+ extrusion. NHE6 and NHE9 are organellar isoforms that can appear transiently on plasma membranes and have been proposed to mediate K+/H+ exchange. We identified NHE6 in a subset of hair bundles; NHE9 was present in all bundles. Heterologous expression of these isoforms in yeast strains lacking endogenous exchangers conferred pH-dependent tolerance to high levels of KCl and NaCl. NHE9 preferred cations in the order K+, Na+ >> Rb+, consistent with the relative efficacies of these ions in promoting pH recovery in hair bundles. Electroneutral K+/H+ exchange, which we propose is performed by NHE9 in hair bundles, exploits the high-K+ endolymph, responds only to pH imbalance across the bundle membrane, is unaffected by the +80 mV endocochlear potential, and uses mechanisms already present in the ear for K+ recycling. This mechanism allows the hair cell to remove H+ generated by Ca2+ pumping without ATP hydrolysis in the cell.
Figures






References
-
- Abacioglu YH, Fouts TR, Laman JD, Claassen E, Pincus SH, Moore JP, Roby CA, Kamin-Lewis R, Lewis GK. Epitope mapping and topology of baculovirus-expressed HIV-1 gp160 determined with a panel of murine monoclonal antibodies. AIDS Res Hum Retroviruses. 1994;10:371–381. - PubMed
-
- Aronson PS. Kinetic properties of the plasma membrane Na+-H+ exchanger. Annu Rev Physiol. 1985;47:545–560. - PubMed
-
- Blair HC, Teitelbaum SL, Ghiselli R, Gluck S. Osteoclastic bone resorption by a polarized vacuolar proton pump. Science. 1989;245:855–857. - PubMed
-
- Bond BR, Ng LL, Schulte BA. Identification of mRNA transcripts and immunohistochemical localization of Na/H exchanger isoforms in gerbil inner ear. Hear Res. 1998;123:1–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous
