Fast cloning inverted repeats for RNA interference
- PMID: 17005926
- PMCID: PMC1624905
- DOI: 10.1261/rna.258406
Fast cloning inverted repeats for RNA interference
Abstract
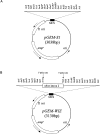
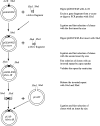
Double-stranded RNA (dsRNA) can induce post-transcriptional gene silencing in a wide variety of organisms. Commonly, inverted repeats are used to produce dsRNA to silence genes of interest. However, cloning inverted repeats still remains a rate-limiting step for widely applying this technique. Here we describe a pGEM-T-based vector, pGEM-WIZ, designed to produce inverted repeats for any Drosophila gene. pGEM-WIZ has a high efficiency in assembling inverted repeats and the repeats in this vector are stable in regular Escherichia coli strains. Furthermore, we have developed a method for rapid selection of clones with an inverted repeat based on size and relative copy number of the vector with or without an insert. This method further eases the cloning process. The inverted repeat cassette assembled in pGEM-WIZ can be easily transferred to commonly available expression vectors suitable for stably expressing inverted repeats in vitro and in vivo.
Figures

References
-
- Bao, S., Cagan, R. Preferential adhesion mediated by Hibris and Roughest regulates morphogenesis and patterning in the Drosophila eye. Dev. Cell. 2005;8:925–935. - PubMed
-
- Brand, A.H., Perrimon, N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. - PubMed
-
- Fire, A., Xu, S., Montgomery, M.K., Kostas, S.A., Driver, S.E., Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans . Nature. 1998;391:806–811. - PubMed
-
- Fortier, E., Belote, J.M. Temperature-dependent gene silencing by an expressed inverted repeat in Drosophila . Genesis. 2000;26:240–244. - PubMed
-
- Holmes, D.S., Quigley, M. A rapid boiling method for the preparation of bacterial plasmids. Anal. Biochem. 1981;114:193–197. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
