Ca2+ sensitivity of regulated cardiac thin filament sliding does not depend on myosin isoform
- PMID: 17008370
- PMCID: PMC1890378
- DOI: 10.1113/jphysiol.2006.120105
Ca2+ sensitivity of regulated cardiac thin filament sliding does not depend on myosin isoform
Abstract
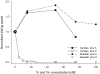
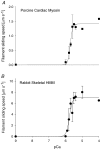
Myosin heavy chain (MHC) isoforms in vertebrate striated muscles are distinguished functionally by differences in chemomechanical kinetics. These kinetic differences may influence the cross-bridge-dependent co-operativity of thin filament Ca(2+) activation. To determine whether Ca(2+) sensitivity of unloaded thin filament sliding depends upon MHC isoform kinetics, we performed in vitro motility assays with rabbit skeletal heavy meromyosin (rsHMM) or porcine cardiac myosin (pcMyosin). Regulated thin filaments were reconstituted with recombinant human cardiac troponin (rhcTn) and alpha-tropomyosin (rhcTm) expressed in Escherichia coli. All three subunits of rhcTn were coexpressed as a functional complex using a novel construct with a glutathione S-transferase (GST) affinity tag at the N-terminus of human cardiac troponin T (hcTnT) and an intervening tobacco etch virus (TEV) protease site that allows purification of rhcTn without denaturation, and removal of the GST tag without proteolysis of rhcTn subunits. Use of this highly purified rhcTn in our motility studies resulted in a clear definition of the regulated motility profile for both fast and slow MHC isoforms. Maximum sliding speed (pCa 5) of regulated thin filaments was roughly fivefold faster with rsHMM compared with pcMyosin, although speed was increased by 1.6- to 1.9-fold for regulated over unregulated actin with both MHC isoforms. The Ca(2+) sensitivity of regulated thin filament sliding speed was unaffected by MHC isoform. Our motility results suggest that the cellular changes in isoform expression that result in regulation of myosin kinetics can occur independently of changes that influence thin filament Ca(2+) sensitivity.
Figures

Similar articles
-
Interaction between troponin and myosin enhances contractile activity of myosin in cardiac muscle.DNA Cell Biol. 2011 Sep;30(9):653-9. doi: 10.1089/dna.2010.1163. Epub 2011 Mar 27. DNA Cell Biol. 2011. PMID: 21438758 Free PMC article.
-
Regulation of force and unloaded sliding speed in single thin filaments: effects of regulatory proteins and calcium.J Physiol. 2000 Apr 1;524 Pt 1(Pt 1):233-43. doi: 10.1111/j.1469-7793.2000.00233.x. J Physiol. 2000. PMID: 10747195 Free PMC article.
-
Calcium regulation of skeletal muscle thin filament motility in vitro.Biophys J. 1997 Mar;72(3):1295-307. doi: 10.1016/S0006-3495(97)78776-9. Biophys J. 1997. PMID: 9138575 Free PMC article.
-
Cardiac contractility: how calcium activates the myofilaments.Naturwissenschaften. 1998 Dec;85(12):575-82. doi: 10.1007/s001140050554. Naturwissenschaften. 1998. PMID: 9871917 Review.
-
Invited Review: pathophysiology of cardiac muscle contraction and relaxation as a result of alterations in thin filament regulation.J Appl Physiol (1985). 2001 Mar;90(3):1125-36. doi: 10.1152/jappl.2001.90.3.1125. J Appl Physiol (1985). 2001. PMID: 11181629 Review.
Cited by
-
Microscopic heat pulses activate cardiac thin filaments.J Gen Physiol. 2019 Jun 3;151(6):860-869. doi: 10.1085/jgp.201812243. Epub 2019 Apr 22. J Gen Physiol. 2019. PMID: 31010810 Free PMC article.
-
Slowed Dynamics of Thin Filament Regulatory Units Reduces Ca2+-Sensitivity of Cardiac Biomechanical Function.Cell Mol Bioeng. 2013 Jun 1;6(2):183-198. doi: 10.1007/s12195-013-0269-8. Cell Mol Bioeng. 2013. PMID: 23833690 Free PMC article.
-
Several cardiomyopathy causing mutations on tropomyosin either destabilize the active state of actomyosin or alter the binding properties of tropomyosin.Biochem Biophys Res Commun. 2011 Mar 4;406(1):74-8. doi: 10.1016/j.bbrc.2011.01.112. Epub 2011 Feb 3. Biochem Biophys Res Commun. 2011. PMID: 21295541 Free PMC article.
-
Removing the regulatory N-terminal domain of cardiac troponin I diminishes incompatibility during bacterial expression.Arch Biochem Biophys. 2007 May 1;461(1):138-45. doi: 10.1016/j.abb.2007.01.011. Epub 2007 Jan 31. Arch Biochem Biophys. 2007. PMID: 17303066 Free PMC article.
-
Interaction between troponin and myosin enhances contractile activity of myosin in cardiac muscle.DNA Cell Biol. 2011 Sep;30(9):653-9. doi: 10.1089/dna.2010.1163. Epub 2011 Mar 27. DNA Cell Biol. 2011. PMID: 21438758 Free PMC article.
References
-
- Alpert NR, Brosseau C, Federico A, Krenz M, Robbins J, Warshaw DM. Molecular mechanics of mouse cardiac myosin isoforms. Am J Physiol Heart Circ Physiol. 2002;283:H1446–H1454. - PubMed
-
- Burkart EM, Sumandea MP, Kobayashi T, Nili M, Martin AF, Homsher E, Solaro RJ. Phosphorylation or glutamic acid substitution at protein kinase C sites on cardiac troponin I differentially depress myofilament tension and shortening velocity. J Biol Chem. 2003;278:11265–11272. - PubMed
-
- Chase PB, Chen Y, Kulin KL, Daniel TL. Viscosity and solute dependence of F-actin translocation by rabbit skeletal heavy meromyosin. Am J Physiol Cell Physiol. 2000;278:C1088–C1098. - PubMed
-
- Chikuni K, Tanabe R, Muroya S, Nakajima I. Comparative sequence analysis of four myosin heavy chain isoforms expressed in porcine skeletal muscles: sequencing and characterization of the porcine myosin heavy chain slow isoform. Anim Sci J. 2002;73:257–262.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

