NMR structural and kinetic characterization of a homeodomain diffusing and hopping on nonspecific DNA
- PMID: 17008406
- PMCID: PMC1622777
- DOI: 10.1073/pnas.0605868103
NMR structural and kinetic characterization of a homeodomain diffusing and hopping on nonspecific DNA
Abstract
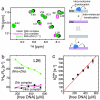
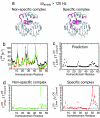
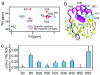
Nonspecific protein-DNA interactions are inherently dynamic and involve both diffusion of the protein along the DNA and hopping of the protein from one DNA molecule or segment to another. Understanding how gene regulatory proteins interact nonspecifically with DNA in terms of both structure and dynamics is challenging because the experimental observables are an ensemble average of many rapidly exchanging states. By using a variety of NMR spectroscopic techniques, including relaxation analysis, paramagnetic relaxation enhancement, and residual dipolar couplings, we have characterized structural and kinetic aspects of the interaction of the HoxD9 homeodomain with a nonspecific, 24-bp DNA duplex in a system in which the protein is not constrained to any particular site. The data reveal that HoxD9 binds to nonspecific DNA with the same binding mode and orientation as that observed in the specific complex. The mobility, however, of Arg side-chains contacting the DNA is increased in the nonspecific complex relative to the specific one. The kinetics of intermolecular translocation between two different nonspecific DNA molecules have also been analyzed and reveal that at high DNA concentrations (such as those present in vivo) direct transfer from one nonspecific complex to another nonspecific DNA molecule occurs without going through the intermediary of free protein. This finding provides a simple mechanism for accelerating the target search in vivo for the specific site in a sea of nonspecific sites by permitting more effective sampling of available DNA sites as the protein jumps from one segment to another.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- von Hippel PH, Berg OG. J Biol Chem. 1989;264:675–678. - PubMed
-
- Lewin B. Genes VII. Oxford: Oxford Univ Press; 2000.
-
- Bruinsma RF. Phys A. 2002;313:211–237.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

