Direct observation of Bin/amphiphysin/Rvs (BAR) domain-induced membrane curvature by means of molecular dynamics simulations
- PMID: 17008407
- PMCID: PMC1622778
- DOI: 10.1073/pnas.0603917103
Direct observation of Bin/amphiphysin/Rvs (BAR) domain-induced membrane curvature by means of molecular dynamics simulations
Abstract
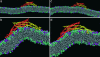
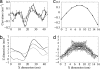
The process of membrane curvature generation by BAR (Bin/amphiphysin/Rvs) domains is thought to involve the plastering of the negatively charged cell membrane to the positively charged concave surface of the BAR domain. Recent work [Peter, B. J., et al. (2004) Science, 303,495-499; Masuda, M., et al. (2006) EMBO J. 25, 2889-2897; and Gallop, J. L., et al. (2006) EMBO J. 25, 2898-2910] has demonstrated the importance of the charged, crescent-shaped surface and the N-terminal amphipathic helices (present in N-BAR domains) for generating membrane curvature. These experiments suggest that curvature is generated by the synergistic action of the N-terminal helices embedding in the lipid bilayer and the charged crescent-shaped dimer acting to "scaffold" membrane curvature. Here, we present atomistic molecular dynamics simulations that directly show membrane binding to the concave face of N-BAR domains, resulting in the generation of local membrane curvature that matches the curvature presented by the BAR domain. These simulations provide direct molecular-scale evidence that BAR domains create curvature by acting as a scaffold, forcing the membrane to locally adopt the intrinsic shape of the BAR domain. We find that BAR domains bind strongly through the maximum curvature surface and, additionally, at an orientation that presents a lesser degree of curvature, thus enabling N-BAR domains to induce a range of local curvatures. Finally, we find that the N-terminal region may play a role in biasing the orientations of N-BAR domains on the membrane surface to those that favor binding to the concave face and subsequent membrane bending.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
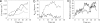

References
-
- Di Paolo G, Sankaranarayanan S, Wenk MR, Daniell L, Perucco E, Caldarone BJ, Flavell R, Picciotto MR, Ryan TA, Cremona O, et al. Neuron. 2002;33:789–804. - PubMed
-
- Zhang B, Zelhof AC. Traffic. 2002;3:452–460. - PubMed
-
- Lee E, Marcucci M, Daniell L, Pypaert M, Weisz OA, Ochoa GC, Farsad K, Wenk MR, De Camilli P. Science. 2002;297:1193–1196. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

