The MRG domain of human MRG15 uses a shallow hydrophobic pocket to interact with the N-terminal region of PAM14
- PMID: 17008723
- PMCID: PMC2242394
- DOI: 10.1110/ps.062397806
The MRG domain of human MRG15 uses a shallow hydrophobic pocket to interact with the N-terminal region of PAM14
Abstract
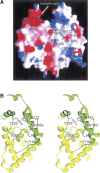
MRG15 is a transcription factor expressed in a variety of human tissues, and its orthologs have been found in many other eukaryotes which constitute the MRG protein family. It plays a vital role in embryonic development and cell proliferation, and is involved in cellular senescence. The C-terminal part of MRG15 forms a conserved MRG domain which is involved in interactions with the tumor suppressor protein retinoblastoma and a nucleoprotein PAM14 during transcriptional regulation. We report here the characterization of the interaction between the MRG domain of human MRG15 and PAM14 using both yeast two-hybrid and in vitro binding assays based on the crystal structure of the MRG domain. The MRG domain is predominantly hydrophobic, and consists of mainly alpha-helices that are arranged in a three-layer sandwich topology. The hydrophobic core is stabilized by interactions among a number of conserved hydrophobic residues. The molecular surface is largely hydrophobic, but contains a few hydrophilic patches. Structure-based site-directed mutagenesis studies identified key residues involved in the binding of PAM14. Structural and biochemical data together demonstrate that the PAM14 binding site is consisted of residues Ile160, Leu168, Val169, Trp172, Tyr235, Val268, and Arg269 of MRG15, which form a shallow hydrophobic pocket to interact with the N-terminal 50 residues of PAM14 through primarily hydrophobic interactions. These results provide the molecular basis for the interaction between the MRG domain and PAM14, and reveal insights into the potential biological function of MRG15 in transcription regulation and chromatin remodeling.
Figures


Similar articles
-
Multipurpose MRG domain involved in cell senescence and proliferation exhibits structural homology to a DNA-interacting domain.Structure. 2006 Jan;14(1):151-8. doi: 10.1016/j.str.2005.08.019. Structure. 2006. PMID: 16407074
-
MRG15 activates the B-myb promoter through formation of a nuclear complex with the retinoblastoma protein and the novel protein PAM14.J Biol Chem. 2001 Oct 19;276(42):39171-8. doi: 10.1074/jbc.M103435200. Epub 2001 Aug 10. J Biol Chem. 2001. PMID: 11500496
-
Interaction of Sedlin with PAM14.J Cell Biochem. 2010 Apr 15;109(6):1129-33. doi: 10.1002/jcb.22491. J Cell Biochem. 2010. PMID: 20108251
-
Structural and functional insights into the epigenetic regulator MRG15.Acta Pharmacol Sin. 2024 May;45(5):879-889. doi: 10.1038/s41401-023-01211-6. Epub 2024 Jan 8. Acta Pharmacol Sin. 2024. PMID: 38191914 Free PMC article. Review.
-
MRGing chromatin dynamics and cellular senescence.Cell Biochem Biophys. 2008;50(3):133-41. doi: 10.1007/s12013-008-9006-7. Epub 2008 Jan 30. Cell Biochem Biophys. 2008. PMID: 18231726 Review.
Cited by
-
Structure of human MRG15 chromo domain and its binding to Lys36-methylated histone H3.Nucleic Acids Res. 2006;34(22):6621-8. doi: 10.1093/nar/gkl989. Epub 2006 Nov 28. Nucleic Acids Res. 2006. PMID: 17135209 Free PMC article.
-
Histone deacetylase 2 (HDAC2) protein-dependent deacetylation of mortality factor 4-like 1 (MORF4L1) protein enhances its homodimerization.J Biol Chem. 2014 Mar 7;289(10):7092-7098. doi: 10.1074/jbc.M113.527507. Epub 2014 Jan 22. J Biol Chem. 2014. PMID: 24451372 Free PMC article.
-
Structural studies on MRG701 chromodomain reveal a novel dimerization interface of MRG proteins in green plants.Protein Cell. 2016 Nov;7(11):792-803. doi: 10.1007/s13238-016-0310-5. Epub 2016 Sep 8. Protein Cell. 2016. PMID: 27638467 Free PMC article.
-
Bromodomain factor 5 is an essential regulator of transcription in Leishmania.Nat Commun. 2022 Jul 13;13(1):4071. doi: 10.1038/s41467-022-31742-1. Nat Commun. 2022. PMID: 35831302 Free PMC article.
-
Characterization of native protein complexes and protein isoform variation using size-fractionation-based quantitative proteomics.Mol Cell Proteomics. 2013 Dec;12(12):3851-73. doi: 10.1074/mcp.M113.032367. Epub 2013 Sep 16. Mol Cell Proteomics. 2013. PMID: 24043423 Free PMC article.
References
-
- Akhtar, A., Zink, D., Becker, P.B. 2000. Chromodomains are protein–RNA interaction modules. Nature 407: 405–409. - PubMed
-
- Bannister, A.J., Zegerman, P., Partridge, J.F., Miska, E.A., Thomas, J.O., Allshire, R.C., Kouzarides, T. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120–124. - PubMed
-
- Bertram, M.J. and Pereira-Smith, O.M. 2001. Conservation of the MORF4 related gene family: Identification of a new chromo domain subfamily and novel protein motif. Gene 266: 111–121. - PubMed
-
- Bertram, M.J., Berube, N.G., Hang-Swanson, X., Ran, Q., Leung, J.K., Bryce, S., Spurgers, K., Bick, R.J., Baldini, A., Ning, Y. et al. 1999. Identification of a gene that reverses the immortal phenotype of a subset of cells and is a member of a novel family of transcription factor-like genes. Mol. Cell. Biol. 19: 1479–1485. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

