Contribution of gene expression to metabolic fluxes in hypermetabolic livers induced through burn injury and cecal ligation and puncture in rats
- PMID: 17009336
- PMCID: PMC3199956
- DOI: 10.1002/bit.21200
Contribution of gene expression to metabolic fluxes in hypermetabolic livers induced through burn injury and cecal ligation and puncture in rats
Abstract
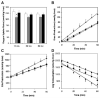
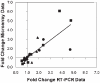
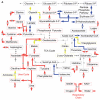
Severe injury activates many stress-related and inflammatory pathways that can lead to a systemic hypermetabolic state. Prior studies using perfused hypermetabolic rat livers have identified intrinsic metabolic flux changes that were not dependent upon the continual presence of elevated stress hormones and substrate loads. We investigated the hypothesis that such changes may be due to persistent alterations in gene expression. A systemic hypermetabolic response was induced in rats by applying a moderate burn injury followed 2 days later by cecum ligation and puncture (CLP) to produce sepsis. Control animals received a sham-burn followed by CLP, or a sham-burn followed by sham-CLP. Two days after CLP, livers were analyzed for gene expression changes using DNA microarrays and for metabolism alterations by ex vivo perfusion coupled with Metabolic Flux Analysis. Burn injury prior to CLP increased fluxes while decreases in gene expression levels were observed. Conversely, CLP alone significantly increased metabolic gene expression, but decreased many of the corresponding metabolic fluxes. Burn injury combined with CLP led to the most dramatic changes, where concurrent changes in fluxes and gene expression levels occurred in about 1/3 of the reactions. The data are consistent with the notion that in this model, burn injury prior to CLP increased fluxes through post-translational mechanisms with little contribution of gene expression, while CLP treatment up-regulated the metabolic machinery by transcriptional mechanisms. Overall, these data show that mRNA changes measured at a single time point by DNA microarray analysis do not reliably predict metabolic flux changes in perfused livers.
(c) 2006 Wiley Periodicals, Inc.
Figures







Similar articles
-
Dynamics of hepatic gene expression profile in a rat cecal ligation and puncture model.J Surg Res. 2012 Aug;176(2):583-600. doi: 10.1016/j.jss.2011.11.1031. Epub 2011 Dec 15. J Surg Res. 2012. PMID: 22381171 Free PMC article.
-
Profiling of dynamic changes in hypermetabolic livers.Biotechnol Bioeng. 2003 Aug 20;83(4):400-15. doi: 10.1002/bit.10682. Biotechnol Bioeng. 2003. PMID: 12800135
-
Proteomic analysis of altered protein expression in skeletal muscle of rats in a hypermetabolic state induced by burn sepsis.Biochem J. 2006 Jul 1;397(1):149-58. doi: 10.1042/BJ20051710. Biochem J. 2006. PMID: 16483253 Free PMC article.
-
The effect of sequential injuries on splanchnic perfusion and eicosanoid release.J Surg Res. 1998 Aug;78(2):148-54. doi: 10.1006/jsre.1997.5250. J Surg Res. 1998. PMID: 9733633
-
Burn injury exacerbates hemodynamic and metabolic responses in rats with polymicrobial sepsis.J Burn Care Res. 2006 Jan-Feb;27(1):50-9. doi: 10.1097/01.bcr.0000192568.77001.b1. J Burn Care Res. 2006. PMID: 16566537
Cited by
-
Metabolic response of perfused livers to various oxygenation conditions.Biotechnol Bioeng. 2011 Dec;108(12):2947-57. doi: 10.1002/bit.23261. Epub 2011 Aug 4. Biotechnol Bioeng. 2011. PMID: 21755498 Free PMC article.
-
Metabolic network-based predictions of toxicant-induced metabolite changes in the laboratory rat.Sci Rep. 2018 Aug 3;8(1):11678. doi: 10.1038/s41598-018-30149-7. Sci Rep. 2018. PMID: 30076366 Free PMC article.
-
Integration of Metabolic Modeling with Gene Co-expression Reveals Transcriptionally Programmed Reactions Explaining Robustness in Mycobacterium tuberculosis.Sci Rep. 2016 Mar 22;6:23440. doi: 10.1038/srep23440. Sci Rep. 2016. PMID: 27000948 Free PMC article.
-
Long-term dynamic profiling of inflammatory mediators in double-hit burn and sepsis animal models.Cytokine. 2012 May;58(2):307-15. doi: 10.1016/j.cyto.2012.01.017. Epub 2012 Mar 7. Cytokine. 2012. PMID: 22402033 Free PMC article.
-
Dynamic epistasis for different alleles of the same gene.Proc Natl Acad Sci U S A. 2012 Jun 26;109(26):10420-5. doi: 10.1073/pnas.1121507109. Epub 2012 Jun 11. Proc Natl Acad Sci U S A. 2012. PMID: 22689976 Free PMC article.
References
-
- Anderson L, Seilhamer J. A comparison of selected mRNA and protein abundances in human liver. Electrophoresis. 1997;18(3-4):533–537. - PubMed
-
- Arai K, Lee K, Berthiaume F, Tompkins RG, Yarmush ML. Intrahe-patic amino acid and glucose metabolism in a D-galactosamine-induced rat liver failure model. Hepatology. 2001;34(2):360–371. - PubMed
-
- Banta S, Yokoyama T, Berthiaume F, Yarmush ML. Quantitative effects of thermal injury and insulin on the metabolism of the skeletal muscle using the perfused rat hindquarter preparation. Biotechnol Bioeng. 2004;88(5):613–629. - PubMed
-
- Banta S, Yokoyama T, Berthiaume F, Yarmush ML. Effects of dehydroepiandrosterone administration on rat hepatic metabolism following thermal injury. J Surg Res. 2005;127(2):93–105. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

