The many faces of protein-protein interactions: A compendium of interface geometry
- PMID: 17009862
- PMCID: PMC1584320
- DOI: 10.1371/journal.pcbi.0020124
The many faces of protein-protein interactions: A compendium of interface geometry
Abstract
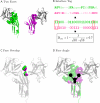
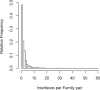
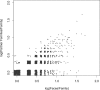
A systematic classification of protein-protein interfaces is a valuable resource for understanding the principles of molecular recognition and for modelling protein complexes. Here, we present a classification of domain interfaces according to their geometry. Our new algorithm uses a hybrid approach of both sequential and structural features. The accuracy is evaluated on a hand-curated dataset of 416 interfaces. Our hybrid procedure achieves 83% precision and 95% recall, which improves the earlier sequence-based method by 5% on both terms. We classify virtually all domain interfaces of known structure, which results in nearly 6,000 distinct types of interfaces. In 40% of the cases, the interacting domain families associate in multiple orientations, suggesting that all the possible binding orientations need to be explored for modelling multidomain proteins and protein complexes. In general, hub proteins are shown to use distinct surface regions (multiple faces) for interactions with different partners. Our classification provides a convenient framework to query genuine gene fusion, which conserves binding orientation in both fused and separate forms. The result suggests that the binding orientations are not conserved in at least one-third of the gene fusion cases detected by a conventional sequence similarity search. We show that any evolutionary analysis on interfaces can be skewed by multiple binding orientations and multiple interaction partners. The taxonomic distribution of interface types suggests that ancient interfaces common to the three major kingdoms of life are enriched by symmetric homodimers. The classification results are online at http://www.scoppi.org.
Conflict of interest statement
Figures

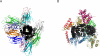




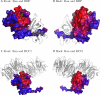
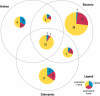
References
-
- Inbar Y, Benyamini H, Nussinov R, Wolfson HJ. Protein structure prediction via combinatorial assembly of sub-structural units. Bioinformatics. 2003;19(Supplement 1):i158–168. - PubMed
-
- Aloy P, Ceulemans H, Stark A, Russell RB. The relationship between sequence and interaction divergence in proteins. J Mol Biol. 2003;332:989–998. - PubMed
-
- Aloy P, Bottcher B, Ceulemans H, Leutwein C, Mellwig C, et al. Structure-based assembly of protein complexes in yeast. Science. 2004;303:2026–2029. - PubMed
-
- Prabu MM, Suguna K, Vijayan M. Variability in quaternary association of proteins with the same tertiary fold: A case study and rationalization involving legume lectins. Proteins. 1999;35:58–69. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

