Targeted molecular dynamics study of C-loop closure and channel gating in nicotinic receptors
- PMID: 17009865
- PMCID: PMC1584325
- DOI: 10.1371/journal.pcbi.0020134
Targeted molecular dynamics study of C-loop closure and channel gating in nicotinic receptors
Abstract
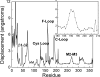
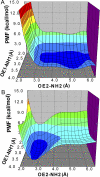
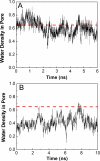
The initial coupling between ligand binding and channel gating in the human alpha7 nicotinic acetylcholine receptor (nAChR) has been investigated with targeted molecular dynamics (TMD) simulation. During the simulation, eight residues at the tip of the C-loop in two alternating subunits were forced to move toward a ligand-bound conformation as captured in the crystallographic structure of acetylcholine binding protein (AChBP) in complex with carbamoylcholine. Comparison of apo- and ligand-bound AChBP structures shows only minor rearrangements distal from the ligand-binding site. In contrast, comparison of apo and TMD simulation structures of the nAChR reveals significant changes toward the bottom of the ligand-binding domain. These structural rearrangements are subsequently translated to the pore domain, leading to a partly open channel within 4 ns of TMD simulation. Furthermore, we confirmed that two highly conserved residue pairs, one located near the ligand-binding pocket (Lys145 and Tyr188), and the other located toward the bottom of the ligand-binding domain (Arg206 and Glu45), are likely to play important roles in coupling agonist binding to channel gating. Overall, our simulations suggest that gating movements of the alpha7 receptor may involve relatively small structural changes within the ligand-binding domain, implying that the gating transition is energy-efficient and can be easily modulated by agonist binding/unbinding.
Conflict of interest statement
Figures









References
-
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, et al. Molecular biology of the cell. 4th edition. New York: Garland Publishing; 2002. 1616
-
- Karlin A. Emerging structure of the nicotinic acetylcholine receptors. Nat Rev Neurosci. 2002;3:102–114. - PubMed
-
- Jensen AA, Frolund B, Liljefors T, Krogsgaard-Larsen P. Neuronal nicotinic acetylcholine receptors: Structural revelations, target identifications, and therapeutic inspirations. J Med Chem. 2005;48:4705–4745. - PubMed
-
- Cassels BK, Bermudez I, Dajas F, Abin-Carriquiry JA, Wonnacott S. From ligand design to therapeutic efficacy: The challenge for nicotinic receptor research. Drug Discov Today. 2005;10:1657–1665. - PubMed
-
- Dajas-Bailador F, Wonnacott S. Nicotinic acetylcholine receptors and the regulation of neuronal signalling. Trends Pharmacol Sci. 2004;25:317–324. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

