Analysis of pleiotropic transcriptional profiles: a case study of DNA gyrase inhibition
- PMID: 17009874
- PMCID: PMC1584274
- DOI: 10.1371/journal.pgen.0020152
Analysis of pleiotropic transcriptional profiles: a case study of DNA gyrase inhibition
Abstract
Genetic and environmental perturbations often result in complex transcriptional responses involving multiple genes and regulons. In order to understand the nature of a response, one has to account for the contribution of the downstream effects to the formation of a response. Such analysis can be carried out within a statistical framework in which the individual effects are independently collected and then combined within a linear model. Here, we modeled the contribution of DNA replication, supercoiling, and repair to the transcriptional response of inhibition of the Escherichia coli gyrase. By representing the gyrase inhibition as a true pleiotropic phenomenon, we were able to demonstrate that: (1) DNA replication is required for the formation of spatial transcriptional domains; (2) the transcriptional response to the gyrase inhibition is coordinated between at least two modules involved in DNA maintenance, relaxation and damage response; (3) the genes whose transcriptional response to the gyrase inhibition does not depend on the main relaxation activity of the cell can be classified on the basis of a GC excess in their upstream and coding sequences; and (4) relaxation by topoisomerase I dominates the transcriptional response, followed by the effects of replication and RecA. We functionally tested the effect of the interaction between relaxation and repair activities, and found support for the model derived from the microarray data. We conclude that modeling compound transcriptional profiles as a combination of downstream transcriptional effects allows for a more realistic, accurate, and meaningful representation of the transcriptional activity of a genome.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures


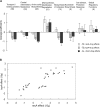
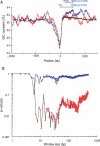
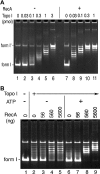
References
-
- Funnell BE, Baker TA, Kornberg A. Complete enzymatic replication of plasmids containing the origin of the Escherichia coli chromosome. J Biol Chem. 1986;261:5616–5624. - PubMed
-
- Steck TR, Drlica K. Bacterial chromosome segregation: Evidence for DNA gyrase involvement in decatenation. Cell. 1984;36:1081–1088. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

