Mutation analysis and characterization of ATR sequence variants in breast cancer cases from high-risk French Canadian breast/ovarian cancer families
- PMID: 17010193
- PMCID: PMC1599749
- DOI: 10.1186/1471-2407-6-230
Mutation analysis and characterization of ATR sequence variants in breast cancer cases from high-risk French Canadian breast/ovarian cancer families
Abstract
Background: Ataxia telangiectasia-mutated and Rad3-related (ATR) is a member of the PIK-related family which plays, along with ATM, a central role in cell-cycle regulation. ATR has been shown to phosphorylate several tumor suppressors like BRCA1, CHEK1 and TP53. ATR appears as a good candidate breast cancer susceptibility gene and the current study was designed to screen for ATR germline mutations potentially involved in breast cancer predisposition.
Methods: ATR direct sequencing was performed using a fluorescent method while widely available programs were used for linkage disequilibrium (LD), haplotype analyses, and tagging SNP (tSNP) identification. Expression analyses were carried out using real-time PCR.
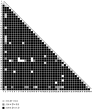
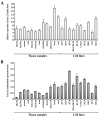
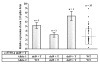
Results: The complete sequence of all exons and flanking intronic sequences were analyzed in DNA samples from 54 individuals affected with breast cancer from non-BRCA1/2 high-risk French Canadian breast/ovarian families. Although no germline mutation has been identified in the coding region, we identified 41 sequence variants, including 16 coding variants, 3 of which are not reported in public databases. SNP haplotypes were established and tSNPs were identified in 73 healthy unrelated French Canadians, providing a valuable tool for further association studies involving the ATR gene, using large cohorts. Our analyses led to the identification of two novel alternative splice transcripts. In contrast to the transcript generated by an alternative splicing site in the intron 41, the one resulting from a deletion of 121 nucleotides in exon 33 is widely expressed, at significant but relatively low levels, in both normal and tumoral cells including normal breast and ovarian tissue.
Conclusion: Although no deleterious mutations were identified in the ATR gene, the current study provides an haplotype analysis of the ATR gene polymorphisms, which allowed the identification of a set of SNPs that could be used as tSNPs for large-scale association studies. In addition, our study led to the characterization of a novel Delta33 splice form, which could generate a putative truncated protein lacking several functional domains. Additional studies in large cohorts and other populations will be needed to further evaluate if common and/or rare ATR sequence variants can be associated with a modest or intermediate breast cancer risk.
Figures




References
-
- Houlston RS, Peto J. Genetics and the common cancers. In: Eeles RA, Easton DF, Ponder BAJ, Eng C, editor. Genetic Predisposition to Cancer. New York: Oxford University Press; 2004. pp. 235–247.
-
- Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, Pukkala E, Skytthe A, Hemminki K. Environmental and heritable factors in the causation of cancer-analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

