Analysis of the prostate cancer cell line LNCaP transcriptome using a sequencing-by-synthesis approach
- PMID: 17010196
- PMCID: PMC1592491
- DOI: 10.1186/1471-2164-7-246
Analysis of the prostate cancer cell line LNCaP transcriptome using a sequencing-by-synthesis approach
Abstract
Background: High throughput sequencing-by-synthesis is an emerging technology that allows the rapid production of millions of bases of data. Although the sequence reads are short, they can readily be used for re-sequencing. By re-sequencing the mRNA products of a cell, one may rapidly discover polymorphisms and splice variants particular to that cell.
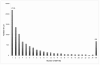
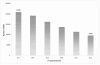
Results: We present the utility of massively parallel sequencing by synthesis for profiling the transcriptome of a human prostate cancer cell-line, LNCaP, that has been treated with the synthetic androgen, R1881. Through the generation of approximately 20 megabases (MB) of EST data, we detect transcription from over 10,000 gene loci, 25 previously undescribed alternative splicing events involving known exons, and over 1,500 high quality single nucleotide discrepancies with the reference human sequence. Further, we map nearly 10,000 ESTs to positions on the genome where no transcription is currently predicted to occur. We also characterize various obstacles with using sequencing by synthesis for transcriptome analysis and propose solutions to these problems.
Conclusion: The use of high-throughput sequencing-by-synthesis methods for transcript profiling allows the specific and sensitive detection of many of a cell's transcripts, and also allows the discovery of high quality base discrepancies, and alternative splice variants. Thus, this technology may provide an effective means of understanding various disease states, discovering novel targets for disease treatment, and discovery of novel transcripts.
Figures
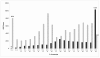

References
-
- Hillier LD, Lennon G, Becker M, Bonaldo MF, Chiapelli B, Chissoe S, Dietrich N, DuBuque T, Favello A, Gish W, Hawkins M, Hultman M, Kucaba T, Lacy M, Le M, Le N, Mardis E, Moore B, Morris M, Parsons J, Prange C, Rifkin L, Rohlfing T, Schellenberg K, Marra M, et al. Generation and analysis of 280,000 human expressed sequence tags. Genome Res. 1996;6:807–828. - PubMed
-
- Hubbard T, Andrews D, Caccamo M, Cameron G, Chen Y, Clamp M, Clarke L, Coates G, Cox T, Cunningham F, Curwen V, Cutts T, Down T, Durbin R, Fernandez-Suarez XM, Gilbert J, Hammond M, Herrero J, Hotz H, Howe K, Iyer V, Jekosch K, Kahari A, Kasprzyk A, Keefe D, Keenan S, Kokocinsci F, London D, Longden I, McVicker G, Melsopp C, Meidl P, Potter S, Proctor G, Rae M, Rios D, Schuster M, Searle S, Severin J, Slater G, Smedley D, Smith J, Spooner W, Stabenau A, Stalker J, Storey R, Trevanion S, Ureta-Vidal A, Vogel J, White S, Woodwark C, Birney E. Ensembl 2005. Nucleic Acids Res. 2005;33:D447–53. doi: 10.1093/nar/gki138. - DOI - PMC - PubMed
-
- Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484–487. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

