Ancient genomic architecture for mammalian olfactory receptor clusters
- PMID: 17010214
- PMCID: PMC1794568
- DOI: 10.1186/gb-2006-7-10-r88
Ancient genomic architecture for mammalian olfactory receptor clusters
Abstract
Background: Mammalian olfactory receptor (OR) genes reside in numerous genomic clusters of up to several dozen genes. Whole-genome sequence alignment nets of five mammals allow their comprehensive comparison, aimed at reconstructing the ancestral olfactory subgenome.
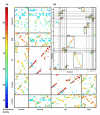
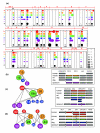
Results: We developed a new and general tool for genome-wide definition of genomic gene clusters conserved in multiple species. Syntenic orthologs, defined as gene pairs showing conservation of both genomic location and coding sequence, were subjected to a graph theory algorithm for discovering CLICs (clusters in conservation). When applied to ORs in five mammals, including the marsupial opossum, more than 90% of the OR genes were found within a framework of 48 multi-species CLICs, invoking a general conservation of gene order and composition. A detailed analysis of individual CLICs revealed multiple differences among species, interpretable through species-specific genomic rearrangements and reflecting complex mammalian evolutionary dynamics. One significant instance involves CLIC #1, which lacks a human member, implying the human-specific deletion of an OR cluster, whose mouse counterpart has been tentatively associated with isovaleric acid odorant detection.
Conclusion: The identified multi-species CLICs demonstrate that most of the mammalian OR clusters have a common ancestry, preceding the split between marsupials and placental mammals. However, only two of these CLICs were capable of incorporating chicken OR genes, parsimoniously implying that all other CLICs emerged subsequent to the avian-mammalian divergence.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

