A novel endonuclease IV post-PCR genotyping system
- PMID: 17012270
- PMCID: PMC1636472
- DOI: 10.1093/nar/gkl679
A novel endonuclease IV post-PCR genotyping system
Abstract
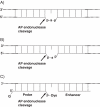
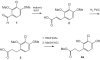
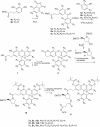
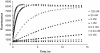
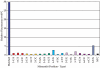
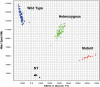
Here we describe a novel endonuclease IV (Endo IV) based assay utilizing a substrate that mimics the abasic lesions that normally occur in double-stranded DNA. The three component substrate is characterized by single-stranded DNA target, an oligonucleotide probe, separated from a helper oligonucleotide by a one base gap. The oligonucleotide probe contains a non-fluorescent quencher at the 5' end and fluorophore attached to the 3' end through a special rigid linker. Fluorescence of the oligonucleotide probe is efficiently quenched by the interaction of terminal dye and quencher when not hybridized. Upon hybridization of the oligonucleotide probe and helper probe to their complementary target, the phosphodiester linkage between the rigid linker and the 3' end of the probe is efficiently cleaved, generating a fluorescent signal. In this study, the use of the Endo IV assay as a post-PCR amplification detection system is demonstrated. High sensitivity and specificity are illustrated using single nucleotide polymorphism detection.
Figures
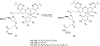





References
-
- Livak K.J. Allelic discrimination using fluorogenic probes and the 5′ nuclease assay. Gen. Anal. 1999;14:143–149. - PubMed
-
- Tang Y.-W., Procop G.W., Persing D.H. Molecular diagnostics of infectious diseases. Clin. Chem. 1997;43:2021–2038. - PubMed
-
- Livak K.J., Flood S.J., Marmaro J., Giusti W., Deetz K. Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl. 1995;4:357–362. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

