Molecular dynamics simulations of human tRNA Lys,3 UUU: the role of modified bases in mRNA recognition
- PMID: 17012271
- PMCID: PMC1636460
- DOI: 10.1093/nar/gkl580
Molecular dynamics simulations of human tRNA Lys,3 UUU: the role of modified bases in mRNA recognition
Abstract
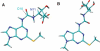
Accuracy in translation of the genetic code into proteins depends upon correct tRNA-mRNA recognition in the context of the ribosome. In human tRNA(Lys,3)UUU three modified bases are present in the anticodon stem-loop--2-methylthio-N6-threonylcarbamoyladenosine at position 37 (ms2t6A37), 5-methoxycarbonylmethyl-2-thiouridine at position 34 (mcm5s2U34) and pseudouridine (psi) at position 39--two of which, ms2t6A37 and mcm5s2U34, are required to achieve wild-type binding activity of wild-type human tRNA(Lys,3)UUU [C. Yarian, M. Marszalek, E. Sochacka, A. Malkiewicz, R. Guenther, A. Miskiewicz and P. F. Agris (2000) Biochemistry, 39, 13390-13395]. Molecular dynamics simulations of nine tRNA anticodon stem-loops with different combinations of nonstandard bases were performed. The wild-type simulation exhibited a canonical anticodon stair-stepped conformation. The ms2t6 modification at position 37 is required for maintenance of this structure and reduces solvent accessibility of U36. Ms2t6A37 generally hydrogen bonds across the loop and may prevent U36 from rotating into solution. A water molecule does coordinate to psi39 most of the simulation time but weakly, as most of the residence lifetimes are <40 ps.
Figures


References
-
- Limbach P.A., Crain P.F., Pomerantz S.C., McCloskey J.A. Structures of posttranscriptionally modified nucleosides from RNA. Biochimie. 1995;77:135–138. - PubMed
-
- Motorin Y., Grosjean H. Appendix 1: chemical structures and classification of posttranscriptionally modified nucleosides in RNA. In: Grosjean H., Benne R., editors. Modification and Editing of RNA. Washington, DC: American Society for Microbiology Press; 1998. pp. 534–549.
-
- Yokoyama S., Nishimura S. Modified nucleosides and codon recognition. In: Soll D., RajBhandary U.L., editors. tRNA: Structure, Biosynthesis and Function. Washington, DC: American Society for Microbiology; 1995. pp. 207–223.

