Quadruplex DNA: sequence, topology and structure
- PMID: 17012276
- PMCID: PMC1636468
- DOI: 10.1093/nar/gkl655
Quadruplex DNA: sequence, topology and structure
Abstract
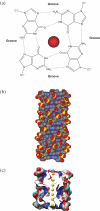
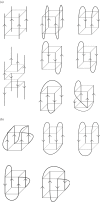
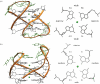
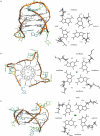
G-quadruplexes are higher-order DNA and RNA structures formed from G-rich sequences that are built around tetrads of hydrogen-bonded guanine bases. Potential quadruplex sequences have been identified in G-rich eukaryotic telomeres, and more recently in non-telomeric genomic DNA, e.g. in nuclease-hypersensitive promoter regions. The natural role and biological validation of these structures is starting to be explored, and there is particular interest in them as targets for therapeutic intervention. This survey focuses on the folding and structural features on quadruplexes formed from telomeric and non-telomeric DNA sequences, and examines fundamental aspects of topology and the emerging relationships with sequence. Emphasis is placed on information from the high-resolution methods of X-ray crystallography and NMR, and their scope and current limitations are discussed. Such information, together with biological insights, will be important for the discovery of drugs targeting quadruplexes from particular genes.
Figures

References
-
- Zimmerman S.B., Cohen G.H., Davies D.R. X-ray fiber diffraction and model-building study of polyguanylic acid and polyinosinic acid. J. Mol. Biol. 1975;92:181–192. - PubMed
-
- Howard F.B., Frazier J., Miles H.T. Stable and metastable forms of poly(G) Biopolymers. 1977;16:791–809. - PubMed
-
- Williamson J.R. G-tetrad structures in telomeric DNA. Ann. Rev. Biophys. Biomol. Struct. 1994;23:703–730. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

