Escherichia coli RNA polymerase recognition of a sigma70-dependent promoter requiring a -35 DNA element and an extended -10 TGn motif
- PMID: 17012380
- PMCID: PMC1698240
- DOI: 10.1128/JB.00853-06
Escherichia coli RNA polymerase recognition of a sigma70-dependent promoter requiring a -35 DNA element and an extended -10 TGn motif
Abstract
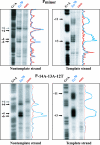
Escherichia coli sigma70-dependent promoters have typically been characterized as either -10/-35 promoters, which have good matches to both the canonical -10 and the -35 sequences or as extended -10 promoters (TGn/-10 promoters), which have the TGn motif and an excellent match to the -10 consensus sequence. We report here an investigation of a promoter, P(minor), that has a nearly perfect match to the -35 sequence and has the TGn motif. However, P(minor) contains an extremely poor sigma70 -10 element. We demonstrate that P(minor) is active both in vivo and in vitro and that mutations in either the -35 or the TGn motif eliminate its activity. Mutation of the TGn motif can be compensated for by mutations that make the -10 element more canonical, thus converting the -35/TGn promoter to a -35/-10 promoter. Potassium permanganate footprinting on the nontemplate and template strands indicates that when polymerase is in a stable (open) complex with P(minor), the DNA is single stranded from positions -11 to +4. We also demonstrate that transcription from P(minor) incorporates nontemplated ribonucleoside triphosphates at the 5' end of the P(minor) transcript, which results in an anomalous assignment for the start site when primer extension analysis is used. P(minor) represents one of the few -35/TGn promoters that have been characterized and serves as a model for investigating functional differences between these promoters and the better-characterized -10/-35 and extended -10 promoters used by E. coli RNA polymerase.
Figures





Similar articles
-
Region 2.5 of the Escherichia coli RNA polymerase sigma70 subunit is responsible for the recognition of the 'extended-10' motif at promoters.EMBO J. 1997 Jul 1;16(13):4034-40. doi: 10.1093/emboj/16.13.4034. EMBO J. 1997. PMID: 9233812 Free PMC article.
-
Interaction of Escherichia coli RNA polymerase σ70 subunit with promoter elements in the context of free σ70, RNA polymerase holoenzyme, and the β'-σ70 complex.J Biol Chem. 2011 Jan 7;286(1):270-9. doi: 10.1074/jbc.M110.174102. Epub 2010 Oct 15. J Biol Chem. 2011. PMID: 20952386 Free PMC article.
-
Domain 1.1 of the sigma(70) subunit of Escherichia coli RNA polymerase modulates the formation of stable polymerase/promoter complexes.J Mol Biol. 2001 Jun 8;309(3):561-72. doi: 10.1006/jmbi.2001.4690. J Mol Biol. 2001. PMID: 11397080
-
The sigma70 family of sigma factors.Genome Biol. 2003;4(1):203. doi: 10.1186/gb-2003-4-1-203. Epub 2003 Jan 3. Genome Biol. 2003. PMID: 12540296 Free PMC article. Review.
-
Analysis of RNA polymerase-promoter complex formation.Methods. 2009 Jan;47(1):13-24. doi: 10.1016/j.ymeth.2008.10.018. Epub 2008 Oct 24. Methods. 2009. PMID: 18952176 Free PMC article. Review.
Cited by
-
Bacteriophage T4 MotA activator and the β-flap tip of RNA polymerase target the same set of σ70 carboxyl-terminal residues.J Biol Chem. 2011 Nov 11;286(45):39290-6. doi: 10.1074/jbc.M111.278762. Epub 2011 Sep 12. J Biol Chem. 2011. PMID: 21911499 Free PMC article.
-
PromoterPredict: sequence-based modelling of Escherichia coli σ70 promoter strength yields logarithmic dependence between promoter strength and sequence.PeerJ. 2018 Nov 7;6:e5862. doi: 10.7717/peerj.5862. eCollection 2018. PeerJ. 2018. PMID: 30425888 Free PMC article.
-
Redefining Escherichia coli σ(70) promoter elements: -15 motif as a complement of the -10 motif.J Bacteriol. 2011 Nov;193(22):6305-14. doi: 10.1128/JB.05947-11. Epub 2011 Sep 9. J Bacteriol. 2011. PMID: 21908667 Free PMC article.
-
Pr is a member of a restricted class of σ70-dependent promoters that lack a recognizable -10 element.Nucleic Acids Res. 2012 Dec;40(22):11308-20. doi: 10.1093/nar/gks934. Epub 2012 Oct 11. Nucleic Acids Res. 2012. PMID: 23066105 Free PMC article.
-
Development of a Metabolite Sensor for High-Throughput Detection of Aldehydes in Escherichia Coli.Front Bioeng Biotechnol. 2018 Aug 23;6:118. doi: 10.3389/fbioe.2018.00118. eCollection 2018. Front Bioeng Biotechnol. 2018. PMID: 30191150 Free PMC article.
References
-
- Browning, D. F., and S. J. Busby. 2004. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2:57-65. - PubMed
-
- Callaci, S., E. Heyduk, and T. Heyduk. 1999. Core RNA polymerase from Escherichia coli induces a major change in the domain arrangement of the sigma 70 subunit. Mol. Cell 3:229-238. - PubMed
-
- Campbell, E. A., O. Muzzin, M. Chlenov, J. L. Sun, C. A. Olson, O. Weinman, M. L. Trester-Zedlitz, and S. A. Darst. 2002. Structure of the bacterial RNA polymerase promoter specificity sigma subunit. Mol. Cell 9:527-539. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

