The cyclin-dependent kinase inhibitor Orysa;KRP1 plays an important role in seed development of rice
- PMID: 17012406
- PMCID: PMC1630760
- DOI: 10.1104/pp.106.087056
The cyclin-dependent kinase inhibitor Orysa;KRP1 plays an important role in seed development of rice
Abstract
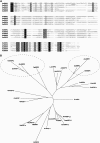
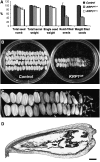
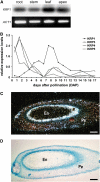
Kip-related proteins (KRPs) play a major role in the regulation of the plant cell cycle. We report the identification of five putative rice (Oryza sativa) proteins that share characteristic motifs with previously described plant KRPs. To investigate the function of KRPs in rice development, we generated transgenic plants overexpressing the Orysa;KRP1 gene. Phenotypic analysis revealed that overexpressed KRP1 reduced cell production during leaf development. The reduced cell production in the leaf meristem was partly compensated by an increased cell size, demonstrating the existence of a compensatory mechanism in monocot species by which growth rate is less reduced than cell production, through cell expansion. Furthermore, Orysa;KRP1 overexpression dramatically reduced seed filling. Sectioning through the overexpressed KRP1 seeds showed that KRP overproduction disturbed the production of endosperm cells. The decrease in the number of fully formed seeds was accompanied by a drop in the endoreduplication of endosperm cells, pointing toward a role of KRP1 in connecting endocycle with endosperm development. Also, spatial and temporal transcript detection in developing seeds suggests that Orysa;KRP1 plays an important role in the exit from the mitotic cell cycle during rice grain formation.
Figures

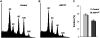

References
-
- Bisbis B, Delmas F, Joubès J, Sicard A, Hernould M, Inzé D, Mouras A, Chevalier C (2006) Cyclin-dependent kinase (CDK) inhibitors regulate the CDK-cyclin complex activities in endoreduplicating cells of developing tomato fruit. J Biol Chem 281: 7374–7383 - PubMed
-
- Boudolf V, Vlieghe K, Beemster GTS, Magyar Z, Torres Acosta JA, Maes S, Van Der Schueren E, Inzé D, De Veylder L (2004) The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2Fa-DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. Plant Cell 16: 2683–2692 - PMC - PubMed
-
- de Almeida Engler J, De Groodt R, Van Montagu M, Engler G (2001) In situ hybridization to mRNA of Arabidopsis tissue sections. Methods 23: 325–334 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

