Evolution and origin of vomeronasal-type odorant receptor gene repertoire in fishes
- PMID: 17014738
- PMCID: PMC1601972
- DOI: 10.1186/1471-2148-6-76
Evolution and origin of vomeronasal-type odorant receptor gene repertoire in fishes
Abstract
Background: In teleost fishes that lack a vomeronasal organ, both main odorant receptors (ORs) and vomeronasal receptors family 2 (V2Rs) are expressed in the olfactory epithelium, and used for perception of water-soluble chemicals. In zebrafish, it is known that both ORs and V2Rs formed multigene families of about a hundred copies. Whereas the contribution of V2Rs in zebrafish to olfaction has been found to be substantially large, the composition and structure of the V2R gene family in other fishes are poorly known, compared with the OR gene family.
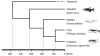
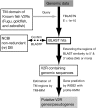
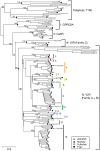
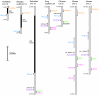
Results: To understand the evolutionary dynamics of V2R genes in fishes, V2R sequences in zebrafish, medaka, fugu, and spotted green pufferfish were identified from their draft genome sequences. There were remarkable differences in the number of intact V2R genes in different species. Most V2R genes in these fishes were tightly clustered in one or two specific chromosomal regions. Phylogenetic analysis revealed that the fish V2R family could be subdivided into 16 subfamilies that had diverged before the separation of the four fishes. Genes in two subfamilies in zebrafish and another subfamily in medaka increased in their number independently, suggesting species-specific evolution in olfaction. Interestingly, the arrangements of V2R genes in the gene clusters were highly conserved among species in the subfamily level. A genomic region of tetrapods corresponding to the region in fishes that contains the V2R cluster was found to have no V2R gene in any species.
Conclusion: Our results have indicated that the evolutionary dynamics of fish V2Rs are characterized by rapid gene turnover and lineage-specific phylogenetic clustering. In addition, the present phylogenetic and comparative genome analyses have shown that the fish V2Rs have expanded after the divergence between teleost and tetrapod lineages. The present identification of the entire V2R repertoire in fishes would provide useful foundation to the future functional and evolutionary studies of fish V2R gene family.
Figures

Similar articles
-
Evolutionary patterns and selective pressures of odorant/pheromone receptor gene families in teleost fishes.PLoS One. 2008;3(12):e4083. doi: 10.1371/journal.pone.0004083. Epub 2008 Dec 31. PLoS One. 2008. PMID: 19116654 Free PMC article.
-
Evolution of vomeronasal-type odorant receptor genes in the zebrafish genome.Gene. 2005 Dec 5;362:19-28. doi: 10.1016/j.gene.2005.07.044. Epub 2005 Oct 14. Gene. 2005. PMID: 16226854
-
Screening the V2R-type putative odorant receptor gene repertoire in bitterling Tanakia lanceolata.Gene. 2009 Jul 15;441(1-2):74-9. doi: 10.1016/j.gene.2008.07.022. Epub 2008 Jul 29. Gene. 2009. PMID: 18706493
-
The molecular evolution of teleost olfactory receptor gene families.Results Probl Cell Differ. 2009;47:37-55. doi: 10.1007/400_2008_11. Results Probl Cell Differ. 2009. PMID: 18956167 Review.
-
Vomeronasal Receptors in Vertebrates and the Evolution of Pheromone Detection.Annu Rev Anim Biosci. 2017 Feb 8;5:353-370. doi: 10.1146/annurev-animal-022516-022801. Epub 2016 Nov 28. Annu Rev Anim Biosci. 2017. PMID: 27912243 Review.
Cited by
-
Co-regulation of a large and rapidly evolving repertoire of odorant receptor genes.BMC Neurosci. 2007 Sep 18;8 Suppl 3(Suppl 3):S2. doi: 10.1186/1471-2202-8-S3-S2. BMC Neurosci. 2007. PMID: 17903278 Free PMC article. Review.
-
Evolutionary patterns and selective pressures of odorant/pheromone receptor gene families in teleost fishes.PLoS One. 2008;3(12):e4083. doi: 10.1371/journal.pone.0004083. Epub 2008 Dec 31. PLoS One. 2008. PMID: 19116654 Free PMC article.
-
The sea lamprey Petromyzon marinus genome reveals the early origin of several chemosensory receptor families in the vertebrate lineage.BMC Evol Biol. 2009 Jul 31;9:180. doi: 10.1186/1471-2148-9-180. BMC Evol Biol. 2009. PMID: 19646260 Free PMC article.
-
Neural circuits mediating olfactory-driven behavior in fish.Front Neural Circuits. 2013 Apr 11;7:62. doi: 10.3389/fncir.2013.00062. eCollection 2013. Front Neural Circuits. 2013. PMID: 23596397 Free PMC article. Review.
-
Remarkable diversity of vomeronasal type 2 receptor (OlfC) genes of basal ray-finned fish and its evolutionary trajectory in jawed vertebrates.Sci Rep. 2022 Apr 19;12(1):6455. doi: 10.1038/s41598-022-10428-0. Sci Rep. 2022. PMID: 35440756 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

