Modelling the checkpoint response to telomere uncapping in budding yeast
- PMID: 17015293
- PMCID: PMC2358953
- DOI: 10.1098/rsif.2006.0148
Modelling the checkpoint response to telomere uncapping in budding yeast
Abstract
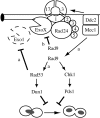
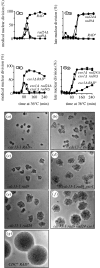
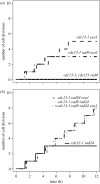
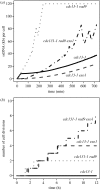
One of the DNA damage-response mechanisms in budding yeast is temporary cell-cycle arrest while DNA repair takes place. The DNA damage response requires the coordinated interaction between DNA repair and checkpoint pathways. Telomeres of budding yeast are capped by the Cdc13 complex. In the temperature-sensitive cdc13-1 strain, telomeres are unprotected over a specific temperature range leading to activation of the DNA damage response and subsequently cell-cycle arrest. Inactivation of cdc13-1 results in the generation of long regions of single-stranded DNA (ssDNA) and is affected by the activity of various checkpoint proteins and nucleases. This paper describes a mathematical model of how uncapped telomeres in budding yeast initiate the checkpoint pathway leading to cell-cycle arrest. The model was encoded in the Systems Biology Markup Language (SBML) and simulated using the stochastic simulation system Biology of Ageing e-Science Integration and Simulation (BASIS). Each simulation follows the time course of one mother cell keeping track of the number of cell divisions, the level of activity of each of the checkpoint proteins, the activity of nucleases and the amount of ssDNA generated. The model can be used to carry out a variety of in silico experiments in which different genes are knocked out and the results of simulation are compared to experimental data. Possible extensions to the model are also discussed.
Figures





References
-
- Foster S.S, Zubko M.K, Guillard S, Lydall D. MRX protects telomeric DNA at uncapped telomeres of budding yeast cdc13-1 mutants. DNA Repair. 2006;5:840–851. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
