Sustained Notch signaling in progenitors is required for sequential emergence of distinct cell lineages during organogenesis
- PMID: 17015435
- PMCID: PMC1578699
- DOI: 10.1101/gad.1444706
Sustained Notch signaling in progenitors is required for sequential emergence of distinct cell lineages during organogenesis
Abstract
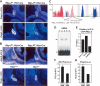
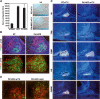
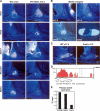
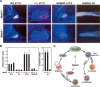
Mammalian organogenesis results from the concerted actions of signaling pathways in progenitor cells that induce a hierarchy of regulated transcription factors critical for organ and cell type determination. Here we demonstrate that sustained Notch activity is required for the temporal maintenance of specific cohorts of proliferating progenitors, which underlies the ability to specify late-arising cell lineages during pituitary organogenesis. Conditional deletion of Rbp-J, which encodes the major mediator of the Notch pathway, leads to premature differentiation of progenitor cells, a phenotype recapitulated by loss of the basic helix-loop-helix (bHLH) factor Hes1, as well as a conversion of the late (Pit1) lineage into the early (corticotrope) lineage. Notch signaling is required for maintaining expression of the tissue-specific paired-like homeodomain transcription factor, Prop1, which is required for generation of the Pit1 lineage. Attenuation of Notch signaling is necessary for terminal differentiation in post-mitotic Pit1+ cells, and the Notch-repressed Pit1 target gene, Math3, is specifically required for maturation and proliferation of the GH-producing somatotrope. Thus, sustained Notch signaling in progenitor cells is required to prevent conversion of the late-arising cell lineages to early-born cell lineages, permitting specification of diverse cell types, a strategy likely to be widely used in mammalian organogenesis.
Figures



References
-
- Artavanis-Tsakonas, S., Rand, M.D., Lake, R.J. Notch signaling: Cell fate control and signal integration in development. Science. 1999;284:770–776. - PubMed
-
- Camper, S.A., Saunders, T.L., Katz, R.W., Reeves, R.H. The Pit-1 transcription factor gene is a candidate for the murine Snell dwarf mutation. Genomics. 1990;8:586–590. - PubMed
-
- Chesnokova, V., Melmed, S. Minireview: Neuro-immuno-endocrine modulation of the hypothalamic-pituitary-adrenal (HPA) axis by gp130 signaling molecules. Endocrinology. 2002;143:1571–1574. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
