Kinesin-13s form rings around microtubules
- PMID: 17015621
- PMCID: PMC2064489
- DOI: 10.1083/jcb.200605194
Kinesin-13s form rings around microtubules
Abstract
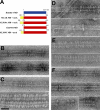
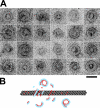
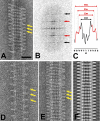
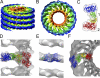
Kinesin is a superfamily of motor proteins that uses the energy of adenosine triphosphate hydrolysis to move and generate force along microtubules. A notable exception to this general description is found in the kinesin-13 family that actively depolymerizes microtubules rather than actively moving along them. This depolymerization activity is important in mitosis during chromosome segregation. It is still not fully clear by which mechanism kinesin-13s depolymerize microtubules. To address this issue, we used electron microscopy to investigate the interaction of kinesin-13s with microtubules. Surprisingly, we found that proteins of the kinesin-13 family form rings and spirals around microtubules. This is the first report of this type of oligomeric structure for any kinesin protein. These rings may allow kinesin-13s to stay at the ends of microtubules during depolymerization.
Figures
References
-
- Carragher, B., M. Whittaker, and R.A. Milligan. 1996. Helical processing using PHOELIX. J. Struct. Biol. 116:107–112. - PubMed
-
- DeRosier, D.J., and P.B. Moore. 1970. Reconstruction of three-dimensional images from electron micrographs of structures with helical symmetry. J. Mol. Biol. 52:355–369. - PubMed
-
- Desai, A., and C.E. Walczak. 2001. Assays for microtubule-destabilizing kinesins. Methods Mol. Biol. 164:109–121. - PubMed
-
- Desai, A., S. Verma, T.J. Mitchison, and C.E. Walczak. 1999. Kin I kinesins are microtubule-destabilizing enzymes. Cell. 96:69–78. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

