BMP1 controls TGFbeta1 activation via cleavage of latent TGFbeta-binding protein
- PMID: 17015622
- PMCID: PMC2064503
- DOI: 10.1083/jcb.200606058
BMP1 controls TGFbeta1 activation via cleavage of latent TGFbeta-binding protein
Abstract
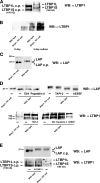
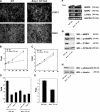
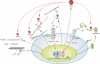
Transforming growth factor beta1 (TGFbeta1), an important regulator of cell behavior, is secreted as a large latent complex (LLC) in which it is bound to its cleaved prodomain (latency-associated peptide [LAP]) and, via LAP, to latent TGFbeta-binding proteins (LTBPs). The latter target LLCs to the extracellular matrix (ECM). Bone morphogenetic protein 1 (BMP1)-like metalloproteinases play key roles in ECM formation, by converting precursors into mature functional proteins, and in morphogenetic patterning, by cleaving the antagonist Chordin to activate BMP2/4. We provide in vitro and in vivo evidence that BMP1 cleaves LTBP1 at two specific sites, thus liberating LLC from ECM and resulting in consequent activation of TGFbeta1 via cleavage of LAP by non-BMP1-like proteinases. In mouse embryo fibroblasts, LAP cleavage is shown to be predominantly matrix metalloproteinase 2 dependent. TGFbeta1 is a potent inducer of ECM formation and of BMP1 expression. Thus, a role for BMP1-like proteinases in TGFbeta1 activation completes a novel fast-forward loop in vertebrate tissue remodeling.
Figures



Similar articles
-
Developmental roles of the BMP1/TLD metalloproteinases.Birth Defects Res C Embryo Today. 2006 Mar;78(1):47-68. doi: 10.1002/bdrc.20060. Birth Defects Res C Embryo Today. 2006. PMID: 16622848 Review.
-
The bone morphogenetic protein 1/Tolloid-like metalloproteinases.Matrix Biol. 2007 Sep;26(7):508-23. doi: 10.1016/j.matbio.2007.05.004. Epub 2007 May 18. Matrix Biol. 2007. PMID: 17560775 Free PMC article. Review.
-
GDF11 forms a bone morphogenetic protein 1-activated latent complex that can modulate nerve growth factor-induced differentiation of PC12 cells.Mol Cell Biol. 2005 Jul;25(14):5846-58. doi: 10.1128/MCB.25.14.5846-5858.2005. Mol Cell Biol. 2005. PMID: 15988002 Free PMC article.
-
Is chordin a long-range- or short-range-acting factor? Roles for BMP1-related metalloproteases in chordin and BMP4 autofeedback loop regulation.Dev Biol. 2000 Jul 1;223(1):120-38. doi: 10.1006/dbio.2000.9740. Dev Biol. 2000. PMID: 10864466
-
Use of Bmp1/Tll1 doubly homozygous null mice and proteomics to identify and validate in vivo substrates of bone morphogenetic protein 1/tolloid-like metalloproteinases.Mol Cell Biol. 2003 Jul;23(13):4428-38. doi: 10.1128/MCB.23.13.4428-4438.2003. Mol Cell Biol. 2003. PMID: 12808086 Free PMC article.
Cited by
-
A SERPINE1-Based Immune Gene Signature Predicts Prognosis and Immunotherapy Response in Gastric Cancer.Pharmaceuticals (Basel). 2022 Nov 14;15(11):1401. doi: 10.3390/ph15111401. Pharmaceuticals (Basel). 2022. PMID: 36422531 Free PMC article.
-
Regulating the cell shift of endothelial cell-like myofibroblasts in pulmonary fibrosis.Eur Respir J. 2023 Jun 8;61(6):2201799. doi: 10.1183/13993003.01799-2022. Print 2023 Jun. Eur Respir J. 2023. PMID: 36758986 Free PMC article.
-
Cellular and molecular mechanisms of thoracic aortic aneurysms.Nat Rev Cardiol. 2009 Dec;6(12):771-86. doi: 10.1038/nrcardio.2009.191. Epub 2009 Nov 3. Nat Rev Cardiol. 2009. PMID: 19884902 Review.
-
Myofibroblast contraction activates latent TGF-beta1 from the extracellular matrix.J Cell Biol. 2007 Dec 17;179(6):1311-23. doi: 10.1083/jcb.200704042. J Cell Biol. 2007. PMID: 18086923 Free PMC article.
-
The role of TGF-β2 and bone morphogenetic proteins in the trabecular meshwork and glaucoma.J Ocul Pharmacol Ther. 2014 Mar-Apr;30(2-3):154-62. doi: 10.1089/jop.2013.0220. Epub 2014 Feb 11. J Ocul Pharmacol Ther. 2014. PMID: 24517218 Free PMC article. Review.
References
-
- Abe, M., J.G. Harpel, C.N. Metz, I. Nunes, D.J. Loskutoff, and D.B. Rifkin. 1994. An assay for transforming growth factor-β using cells transfected with a plasminogen activator inhibitor-1 promoter-luciferase construct. Anal. Biochem. 216:276–284. - PubMed
-
- Annes, J.P., J.S. Munger, and D.B. Rifkin. 2003. Making sense of latent TGF β activation. J. Cell Sci. 116:217–224. - PubMed
-
- Bertolino, P., M. Deckers, F. Lebrin, and P. ten Dijke. 2005. Transforming growth factor-β signal transduction in angiogenesis and vascular disorders. Chest. 128:585S–590S. - PubMed
-
- Blader, P., S. Rastegar, N. Fischer, and U. Strahle. 1997. Cleavage of the BMP-4 antagonist chordin by zebrafish tolloid. Science. 278:1937–1940. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

