Comparative phylogenomics of Clostridium difficile reveals clade specificity and microevolution of hypervirulent strains
- PMID: 17015669
- PMCID: PMC1636221
- DOI: 10.1128/JB.00664-06
Comparative phylogenomics of Clostridium difficile reveals clade specificity and microevolution of hypervirulent strains
Abstract
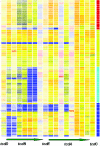
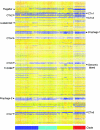
Clostridium difficile is the most frequent cause of nosocomial diarrhea worldwide, and recent reports suggested the emergence of a hypervirulent strain in North America and Europe. In this study, we applied comparative phylogenomics (whole-genome comparisons using DNA microarrays combined with Bayesian phylogenies) to model the phylogeny of C. difficile, including 75 diverse isolates comprising hypervirulent, toxin-variable, and animal strains. The analysis identified four distinct statistically supported clusters comprising a hypervirulent clade, a toxin A(-) B(+) clade, and two clades with human and animal isolates. Genetic differences among clades revealed several genetic islands relating to virulence and niche adaptation, including antibiotic resistance, motility, adhesion, and enteric metabolism. Only 19.7% of genes were shared by all strains, confirming that this enteric species readily undergoes genetic exchange. This study has provided insight into the possible origins of C. difficile and its evolution that may have implications in disease control strategies.
Figures


References
-
- Brazier, J. S. 2001. Typing of Clostridium difficile. Clin. Microbiol. Infect. 7:428-431. - PubMed
-
- Chaves-Olarte, E., P. Low, E. Freer, T. Norlin, M. Weidmann, C. von Eichel-Streiber, and M. Thelestam. 1999. A novel cytotoxin from Clostridium difficile serogroup F is a functional hybrid between two other large clostridial cytotoxins. J. Biol. Chem. 274:11046-11052. - PubMed
-
- Clabots, C. R., S. Johnson, K. M. Bettin, P. A. Mathie, M. E. Mulligan, D. R. Schaberg, L. R. Peterson, and D. N. Gerding. 1993. Development of a rapid and efficient restriction endonuclease analysis typing system for Clostridium difficile and correlation with other typing systems. J. Clin. Microbiol. 31:1870-1875. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

