Evolution of sensory complexity recorded in a myxobacterial genome
- PMID: 17015832
- PMCID: PMC1622800
- DOI: 10.1073/pnas.0607335103
Evolution of sensory complexity recorded in a myxobacterial genome
Erratum in
- Proc Natl Acad Sci U S A. 2006 Dec 19;103(51):19605. Eisen, J [corrected to Eisen, J A]
Abstract
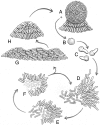
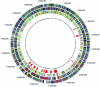
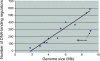
Myxobacteria are single-celled, but social, eubacterial predators. Upon starvation they build multicellular fruiting bodies using a developmental program that progressively changes the pattern of cell movement and the repertoire of genes expressed. Development terminates with spore differentiation and is coordinated by both diffusible and cell-bound signals. The growth and development of Myxococcus xanthus is regulated by the integration of multiple signals from outside the cells with physiological signals from within. A collection of M. xanthus cells behaves, in many respects, like a multicellular organism. For these reasons M. xanthus offers unparalleled access to a regulatory network that controls development and that organizes cell movement on surfaces. The genome of M. xanthus is large (9.14 Mb), considerably larger than the other sequenced delta-proteobacteria. We suggest that gene duplication and divergence were major contributors to genomic expansion from its progenitor. More than 1,500 duplications specific to the myxobacterial lineage were identified, representing >15% of the total genes. Genes were not duplicated at random; rather, genes for cell-cell signaling, small molecule sensing, and integrative transcription control were amplified selectively. Families of genes encoding the production of secondary metabolites are overrepresented in the genome but may have been received by horizontal gene transfer and are likely to be important for predation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Kaiser D. Annu Rev Microbiol. 2004;58:75–98. - PubMed
-
- Dworkin M, Kaiser D, editors. Myxobacteria II. Washington, DC: Am Soc Microbiol; 1993.
-
- Reichenbach H. In: Myxobacteria II. Dworkin M, Kaiser D, editors. Washington, DC: Am Soc Microbiol; 1993. pp. 13–62.
-
- Lynch M, Conery JS. Science. 2003;302:1401–1404. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

