Global gene expression of Prochlorococcus ecotypes in response to changes in nitrogen availability
- PMID: 17016519
- PMCID: PMC1682016
- DOI: 10.1038/msb4100087
Global gene expression of Prochlorococcus ecotypes in response to changes in nitrogen availability
Abstract
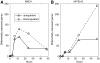
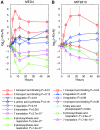
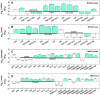
Nitrogen (N) often limits biological productivity in the oceanic gyres where Prochlorococcus is the most abundant photosynthetic organism. The Prochlorococcus community is composed of strains, such as MED4 and MIT9313, that have different N utilization capabilities and that belong to ecotypes with different depth distributions. An interstrain comparison of how Prochlorococcus responds to changes in ambient nitrogen is thus central to understanding its ecology. We quantified changes in MED4 and MIT9313 global mRNA expression, chlorophyll fluorescence, and photosystem II photochemical efficiency (Fv/Fm) along a time series of increasing N starvation. In addition, the global expression of both strains growing in ammonium-replete medium was compared to expression during growth on alternative N sources. There were interstrain similarities in N regulation such as the activation of a putative NtcA regulon during N stress. There were also important differences between the strains such as in the expression patterns of carbon metabolism genes, suggesting that the two strains integrate N and C metabolism in fundamentally different ways.
Figures



References
-
- Allen MM (1984) Cyanobacterial cell inclusions. Annu Rev Microbiol 38: 1–25 - PubMed
-
- Baldi P, Long AD (2001) A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics 17: 509–519 - PubMed
-
- Bertilsson S, Berglund O, Karl DM, Chisholm SW (2003) Elemental composition of marine Prochlorococcus and Synechococcus: implications for the ecological stoichiometry of the sea. Limnol Oceanogr 48: 1721–1731
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

