Targeting of mutant hogg1 in mammalian mitochondria and nucleus: effect on cellular survival upon oxidative stress
- PMID: 17018150
- PMCID: PMC1633743
- DOI: 10.1186/1471-2407-6-235
Targeting of mutant hogg1 in mammalian mitochondria and nucleus: effect on cellular survival upon oxidative stress
Abstract
Background: Oxidative damage to mitochondrial DNA has been implicated as a causative factor in a wide variety of degenerative diseases, aging and cancer. The modified guanine, 7,8-dihydro-8-oxoguanine (also known as 8-hydroxyguanine) is one of the major oxidized bases generated in DNA by reactive oxygen species and has gained most of the attention in recent years as a marker of oxidative DNA injury and its suspected role in the initiation of carcinogenesis. 8-hydroxyguanine is removed by hOgg1, a DNA glycosylase/AP lyase involved in the base excision repair pathway.
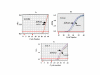
Methods: We over-expressed wild type and R229Q mutant hOGG1 in the nucleus and mitochondria of cells lacking mitochondrial hOGG1 expression through an expression vector containing nuclear and mitochondrial targeting sequence respectively. We used quantitative real time PCR to analyze mtDNA integrity after exposure to oxidative damaging agents, in cells transfected with or without mitochondrially-targeted mutant hogg1.
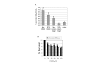
Result: Over-expression of wild type hOgg1 in both nucleus and mitochondria resulted in increased cellular survival when compared to vector or mutant over-expression of hOGG1. Interestingly, mitochondrially-targeted mutant hogg1 resulted in more cell death than nuclear targeted mutant hogg1 upon exposure of cells to oxidative damage. Additional we examined mitochondrial DNA integrity after oxidative damage exposure using real-time quantitative PCR. The presence of mutant hogg1 in the mitochondria resulted in reduced mitochondrial DNA integrity when compared to the wild type. Our work indicates that the R229Q hOGG1 mutation failed to protect cells from oxidative damage and that such mutations in cancer may be more detrimental to cellular survival when present in the mitochondria than in the nucleus.
Conclusion: These findings suggest that deficiencies in hOGG1, especially in the mitochondria may lead to reduced mitochondrial DNA integrity, consequently resulting in decreased cell viability.
Figures



References
-
- Wallace DC, Lott MT, Shoffner JM, Ballinger S. Mitochondrial DNA mutations in epilepsy and neurological disease. Epilepsia. 1994;35 Suppl 1:S43–50. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

