Design of a combinatorial DNA microarray for protein-DNA interaction studies
- PMID: 17018151
- PMCID: PMC1635571
- DOI: 10.1186/1471-2105-7-429
Design of a combinatorial DNA microarray for protein-DNA interaction studies
Abstract
Background: Discovery of precise specificity of transcription factors is an important step on the way to understanding the complex mechanisms of gene regulation in eukaryotes. Recently, double-stranded protein-binding microarrays were developed as a potentially scalable approach to tackle transcription factor binding site identification.
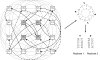
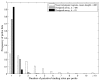
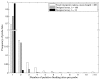
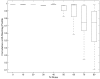
Results: Here we present an algorithmic approach to experimental design of a microarray that allows for testing full specificity of a transcription factor binding to all possible DNA binding sites of a given length, with optimally efficient use of the array. This design is universal, works for any factor that binds a sequence motif and is not species-specific. Furthermore, simulation results show that data produced with the designed arrays is easier to analyze and would result in more precise identification of binding sites.
Conclusion: In this study, we present a design of a double stranded DNA microarray for protein-DNA interaction studies and show that our algorithm allows optimally efficient use of the arrays for this purpose. We believe such a design will prove useful for transcription factor binding site identification and other biological problems.
Figures

References
-
- Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, Yuh CH, Minokawa T, Amore G, Hinman V, Arenas-Mena C, Otim O, Brown CT, Livi CB, Lee PY, Revilla R, Rust AG, Pan Z, Schilstra MJ, Clarke PJ, Arnone MI, Rowen L, Cameron RA, McClay DR, Hood L, Bolouri H. A genomic regulatory network for development. Science. 2002;295:1669–1678. doi: 10.1126/science.1069883. - DOI - PubMed
-
- Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, Zeitlinger J, Jennings EG, Murray HL, Gordon DB, Ren B, Wyrick JJ, Tagne JB, Volkert TL, Fraenkel E, Gifford DK, Young RA. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. - DOI - PubMed
-
- Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, Jennings EG, Zeitlinger J, Pokholok DK, Kellis M, Rolfe PA, Takusagawa KT, Lander ES, Gifford DK, Fraenkel E, Young RA. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. - DOI - PMC - PubMed

