Arrestins: ubiquitous regulators of cellular signaling pathways
- PMID: 17020596
- PMCID: PMC1794542
- DOI: 10.1186/gb-2006-7-9-236
Arrestins: ubiquitous regulators of cellular signaling pathways
Abstract
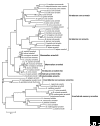
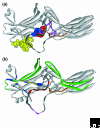
In vertebrates, the arrestins are a family of four proteins that regulate the signaling and trafficking of hundreds of different G-protein-coupled receptors (GPCRs). Arrestin homologs are also found in insects, protochordates and nematodes. Fungi and protists have related proteins but do not have true arrestins. Structural information is available only for free (unbound) vertebrate arrestins, and shows that the conserved overall fold is elongated and composed of two domains, with the core of each domain consisting of a seven-stranded beta-sandwich. Two main intramolecular interactions keep the two domains in the correct relative orientation, but both of these interactions are destabilized in the process of receptor binding, suggesting that the conformation of bound arrestin is quite different. As well as binding to hundreds of GPCR subtypes, arrestins interact with other classes of membrane receptors and more than 20 surprisingly diverse types of soluble signaling protein. Arrestins thus serve as ubiquitous signaling regulators in the cytoplasm and nucleus.
Figures
References
-
- Yamaki K, Tsuda M, Kikuchi T, Chen KH, Huang KP, Shinohara T. Structural organization of the human S-antigen gene. cDNA, amino acid, intron, exon, promoter, in vitro transcription, retina, and pineal gland. J Biol Chem. 1990;265:20757–20762. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

