Complete plastid genome sequences of Drimys, Liriodendron, and Piper: implications for the phylogenetic relationships of magnoliids
- PMID: 17020608
- PMCID: PMC1626487
- DOI: 10.1186/1471-2148-6-77
Complete plastid genome sequences of Drimys, Liriodendron, and Piper: implications for the phylogenetic relationships of magnoliids
Abstract
Background: The magnoliids with four orders, 19 families, and 8,500 species represent one of the largest clades of early diverging angiosperms. Although several recent angiosperm phylogenetic analyses supported the monophyly of magnoliids and suggested relationships among the orders, the limited number of genes examined resulted in only weak support, and these issues remain controversial. Furthermore, considerable incongruence resulted in phylogenetic reconstructions supporting three different sets of relationships among magnoliids and the two large angiosperm clades, monocots and eudicots. We sequenced the plastid genomes of three magnoliids, Drimys (Canellales), Liriodendron (Magnoliales), and Piper (Piperales), and used these data in combination with 32 other angiosperm plastid genomes to assess phylogenetic relationships among magnoliids and to examine patterns of variation of GC content.
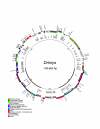
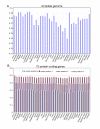
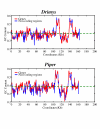
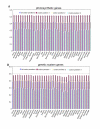
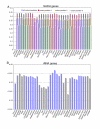
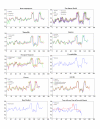
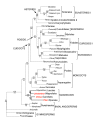
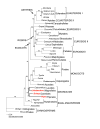
Results: The Drimys, Liriodendron, and Piper plastid genomes are very similar in size at 160,604, 159,886 bp, and 160,624 bp, respectively. Gene content and order are nearly identical to many other unrearranged angiosperm plastid genomes, including Calycanthus, the other published magnoliid genome. Overall GC content ranges from 34-39%, and coding regions have a substantially higher GC content than non-coding regions. Among protein-coding genes, GC content varies by codon position with 1st codon > 2nd codon > 3rd codon, and it varies by functional group with photosynthetic genes having the highest percentage and NADH genes the lowest. Phylogenetic analyses using parsimony and likelihood methods and sequences of 61 protein-coding genes provided strong support for the monophyly of magnoliids and two strongly supported groups were identified, the Canellales/Piperales and the Laurales/Magnoliales. Strong support is reported for monocots and eudicots as sister clades with magnoliids diverging before the monocot-eudicot split. The trees also provided moderate or strong support for the position of Amborella as sister to a clade including all other angiosperms.
Conclusion: Evolutionary comparisons of three new magnoliid plastid genome sequences, combined with other published angiosperm genomes, confirm that GC content is unevenly distributed across the genome by location, codon position, and functional group. Furthermore, phylogenetic analyses provide the strongest support so far for the hypothesis that the magnoliids are sister to a large clade that includes both monocots and eudicots.
Figures

References
-
- Darwin C. Letter to J. D. Hooker. In: Darwin F, Seward AC, editor. More letters of Charles Darwin. Vol. 2. London: John Murray; 1903.
-
- Barkman TJ, Chenery G, McNeal JR, Lyons-Weiler J, Ellisens WJ, Moore G, Wolfe AD, dePamphilis CW. Independent and combined analyses of sequences from all three genomic compartments converge on the root of flowering plant phylogeny. Proc Natl Acad Sci USA. 2000;97:13166–13171. doi: 10.1073/pnas.220427497. - DOI - PMC - PubMed
-
- Doyle JA, Endress PK. Morphological phylogenetic analysis of basal angiosperms: Comparison and combination with molecular data. Int J Plt Sci. 2000;161:S121–S153. doi: 10.1086/317578. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

