Identification of multiple transcription initiation, polyadenylation, and splice sites in the Drosophila melanogaster TART family of telomeric retrotransposons
- PMID: 17020919
- PMCID: PMC1636488
- DOI: 10.1093/nar/gkl709
Identification of multiple transcription initiation, polyadenylation, and splice sites in the Drosophila melanogaster TART family of telomeric retrotransposons
Abstract
The Drosophila non-long terminal repeat (non-LTR) retrotransposons TART and HeT-A specifically retrotranspose to chromosome ends to maintain Drosophila telomeric DNA. Relatively little is known, though, about the regulation of their expression and their retrotransposition to telomeres. We have used rapid amplification of cDNA ends (RACE) to identify multiple transcription initiation and polyadenylation sites for sense and antisense transcripts of three subfamilies of TART elements in Drosophila melanogaster. These results are consistent with the production of an array of TART transcripts. In contrast to other Drosophila non-LTR elements, a major initiation site for sense transcripts was mapped near the 3' end of the TART 5'-untranslated region (5'-UTR), rather than at the start of the 5'-UTR. A sequence overlapping this sense start site contains a good match to an initiator consensus for the transcription start sites of Drosophila LTR retrotransposons. Interestingly, analysis of 5' RACE products for antisense transcripts and the GenBank EST database revealed that TART antisense transcripts contain multiple introns. Our results highlight differences between transcription of TART and of other Drosophila non-LTR elements and they provide a foundation for testing the relationship between exceptional aspects of TART transcription and TART's specialized role at telomeres.
Figures

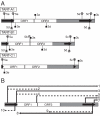
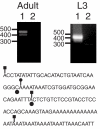


References
-
- Biessmann H., Mason J.M. The unusual telomeres of Drosophila. Trends Genet. 1995;11:58–62. - PubMed
-
- Pardue M.L., DeBaryshe P.G. Retrotransposons provide an evolutionarily robust non-telomerase mechanism to maintain telomeres. Annu. Rev. Genet. 2003;37:485–511. - PubMed
-
- Levis R.W., Ganesan R., Houtchens K., Tolar L.A., Sheen F.M. Transposons in place of telomeric repeats at a Drosophila telomere. Cell. 1993;75:1083–1093. - PubMed
-
- McEachern M.J., Krauskopf A., Blackburn E.H. Telomeres and their control. Annu. Rev. Genet. 2000;34:331–358. - PubMed
-
- Abad J.P., De Pablos B., Osoegawa K., De Jong P.J., Martin-Gallardo A., Villasante A. TAHRE, a novel telomeric retrotransposon from Drosophila melanogaster, reveals the origin of Drosophila telomeres. Mol. Biol. Evol. 2004;21:1620–1624. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

