Analysis of the cytosolic domains of the hepatitis B virus envelope proteins for their function in viral particle assembly and infectivity
- PMID: 17020942
- PMCID: PMC1676254
- DOI: 10.1128/JVI.00621-06
Analysis of the cytosolic domains of the hepatitis B virus envelope proteins for their function in viral particle assembly and infectivity
Abstract
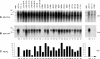
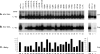
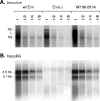
The hepatitis B virus (HBV) envelope proteins have the ability to assemble three types of viral particles, (i) the empty subviral particles (SVPs), (ii) the mature HBV virions, and (iii) the hepatitis delta virus (HDV) particles, in cells that are coinfected with HBV and HDV. To gain insight into the function of the HBV envelope proteins in morphogenesis of HBV or HDV virions, we have investigated subdomains of the envelope proteins that have been shown or predicted to lie at the cytosolic face of the endoplasmic reticulum membrane during synthesis, a position prone to interaction with the inner core structure. These domains, referred to here as cytosolic loops I and II (CYL-I and -II, respectively), were subjected to mutagenesis. The mutations were introduced in the three HBV envelope proteins, designated small, middle, and large (S-HBsAg, M-HBsAg, and L-HBsAg, respectively). The mutants were expressed in HuH-7 cells to evaluate their capacity for self-assembly and formation of HBV or HDV virions when HBV nucleocapsid or HDV ribonucleoprotein, respectively, was provided. We found that SVP-competent CYL-I mutations between positions 23 and 78 of the S domain were permissive to HBV or HDV virion assembly. One mutation (P29A) was permissive for synthesis of the S- and M-HBsAg but adversely affected the synthesis or stability of L-HBsAg, thereby preventing the assembly of HBV virions. Furthermore, using an in vitro infection assay based on the HepaRG cells and the HDV model, we have shown that particles coated with envelope proteins bearing CYL-I mutations were fully infectious, hence indicating the absence of an infectivity determinant in this region. Finally, we demonstrated that the tryptophan residues at positions 196, 199, and 201 in CYL-II, which were shown to exert a matrix function for assembly of HDV particles (I. Komla-Soukha and C. Sureau, J. Virol. 80:4648-4655, 2006), were dispensable for both assembly and infectivity of HBV virions.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

