Kaposi's sarcoma-associated herpesvirus ori-Lyt-dependent DNA replication: dual role of replication and transcription activator
- PMID: 17020951
- PMCID: PMC1676287
- DOI: 10.1128/JVI.00990-06
Kaposi's sarcoma-associated herpesvirus ori-Lyt-dependent DNA replication: dual role of replication and transcription activator
Abstract
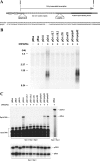
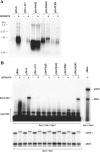
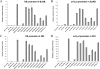
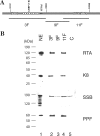
Lytic replication of Kaposi's sarcoma-associated herpesvirus (KSHV) is essential for viral propagation and pathogenicity. In Kaposi's sarcoma lesions, constant lytic replication plays a role in sustaining the population of latently infected cells that otherwise are quickly lost by segregation of latent viral episomes as spindle cells divide. Lytic DNA replication initiates from an origin (ori-Lyt) and requires trans-acting elements. Two functional ori-Lyts have been identified in the KSHV genome. Some cis-acting and trans-acting elements for ori-Lyt-dependent DNA replication have been found. Among these, K8 binding sites, a cluster of C/EBP binding motifs, and a replication and transcription activator (RTA) responsive element (RRE) are crucial cis-acting elements. Binding of K8 and RTA proteins to these motifs in ori-Lyt DNA was demonstrated to be absolutely essential for DNA replication. In the present study, functional roles of RTA in ori-Lyt-dependent DNA replication have been investigated. Two distinct functions of RTA were revealed. First, RTA activates an ori-Lyt promoter and initiates transcription across GC-rich tandem repeats. This RTA-mediated transcription is indispensable for DNA replication. Second, RTA is a component of the replication compartment, where RTA interacts with prereplication complexes composed of at least six core machinery proteins and K8. The prereplication complexes are recruited to ori-Lyt DNA through RTA, which interacts with the RRE, as well as K8, which binds to a cluster of C/EBP binding motifs with the aid of C/EBP alpha. The revelation of these two functions of RTA, together with its role in initiation of a transcriptional cascade that leads to transcription of all viral lytic genes, shows that RTA is a critical initiator and regulator of KSHV lytic DNA replication and viral propagation.
Figures






References
-
- AuCoin, D. P., K. S. Colletti, S. A. Cei, I. Papouskova, M. Tarrant, and G. S. Pari. 2004. Amplification of the Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 lytic origin of DNA replication is dependent upon a cis-acting AT-rich region and an ORF50 response element and the trans-acting factors ORF50 (K-Rta) and K8 (K-bZIP). Virology 318:542-555. - PubMed
-
- Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641-644. - PubMed
-
- Cesarman, E., P. S. Moore, P. H. Rao, G. Inghirami, D. M. Knowles, and Y. Chang. 1995. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi's sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood 86:2708-2714. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

