Species-specific identification of Leptospiraceae by 16S rRNA gene sequencing
- PMID: 17021075
- PMCID: PMC1594759
- DOI: 10.1128/JCM.00670-06
Species-specific identification of Leptospiraceae by 16S rRNA gene sequencing
Abstract
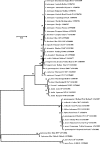
The genus Leptospira is classified into 13 named species and 4 genomospecies based upon DNA-DNA reassociation studies. Phenotypic tests are unable to distinguish between species of Leptospira, and there is a need for a simplified molecular approach to the identification of leptospires. 16S rRNA gene sequences are potentially useful for species identification of Leptospira, but there are a large number of sequences of various lengths and quality in the public databases. 16S rRNA gene sequences of near full length and bidirectional high redundancy were determined for all type strains of the species of the Leptospiraceae. Three clades were identified within the genus Leptospira, composed of pathogenic species, nonpathogenic species, and another clade of undetermined pathogenicity with intermediate 16S rRNA gene sequence relatedness. All type strains could be identified by 16S rRNA gene sequences, but within both pathogenic and nonpathogenic clades as few as two or three base pairs separated some species. Sequences within the nonpathogenic clade were more similar, and in most cases < or =10 bp distinguished these species. These sequences provide a reference standard for identification of Leptospira species and confirm previously established relationships within the genus. 16S rRNA gene sequencing is a powerful method for identification in the clinical laboratory and offers a simplified approach to the identification of Leptospira species.
Figures

References
-
- Brenner, D. J., A. F. Kaufmann, K. R. Sulzer, A. G. Steigerwalt, F. C. Rogers, and R. S. Weyant. 1999. Further determination of DNA relatedness between serogroups and serovars in the family Leptospiraceae with a proposal for Leptospira alexanderi sp. nov. and four new Leptospira genomospecies. Int. J. Syst. Bacteriol. 49:839-858. - PubMed
-
- Busse, H. J., E. B. Denner, and W. Lubitz. 1996. Classification and identification of bacteria: current approaches to an old problem. J. Biotechnol. 47:3-38. - PubMed
-
- de la Peña-Moctezuma, A., D. M. Bulach, T. Kalambaheti, and B. Adler. 1999. Comparative analysis of the LPS biosynthetic loci of the genetic subtypes of serovar Hardjo: Leptospira interrogans subtype Hardjoprajitno and Leptospira borgpetersenii subtype Hardjobovis. FEMS Microbiol. Lett. 177:319-326. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

