M-GCAT: interactively and efficiently constructing large-scale multiple genome comparison frameworks in closely related species
- PMID: 17022809
- PMCID: PMC1629028
- DOI: 10.1186/1471-2105-7-433
M-GCAT: interactively and efficiently constructing large-scale multiple genome comparison frameworks in closely related species
Abstract
Background: Due to recent advances in whole genome shotgun sequencing and assembly technologies, the financial cost of decoding an organism's DNA has been drastically reduced, resulting in a recent explosion of genomic sequencing projects. This increase in related genomic data will allow for in depth studies of evolution in closely related species through multiple whole genome comparisons.
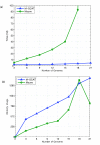
Results: To facilitate such comparisons, we present an interactive multiple genome comparison and alignment tool, M-GCAT, that can efficiently construct multiple genome comparison frameworks in closely related species. M-GCAT is able to compare and identify highly conserved regions in up to 20 closely related bacterial species in minutes on a standard computer, and as many as 90 (containing 75 cloned genomes from a set of 15 published enterobacterial genomes) in an hour. M-GCAT also incorporates a novel comparative genomics data visualization interface allowing the user to globally and locally examine and inspect the conserved regions and gene annotations.
Conclusion: M-GCAT is an interactive comparative genomics tool well suited for quickly generating multiple genome comparisons frameworks and alignments among closely related species. M-GCAT is freely available for download for academic and non-commercial use at: http://alggen.lsi.upc.es/recerca/align/mgcat/intro-mgcat.html.
Figures



References
-
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer MLI, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, Mcdade KE, Mckenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

