Gene-expression signature of benign monoclonal gammopathy evident in multiple myeloma is linked to good prognosis
- PMID: 17023574
- PMCID: PMC1794073
- DOI: 10.1182/blood-2006-07-037077
Gene-expression signature of benign monoclonal gammopathy evident in multiple myeloma is linked to good prognosis
Abstract
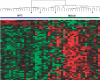
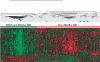
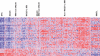
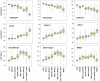
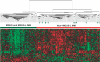
Monoclonal gammopathy of undetermined significance (MGUS) can progress to multiple myeloma (MM). Although these diseases share many of the same genetic features, it is still unclear whether global gene-expression profiling might identify prior genomic signatures that distinguish them. Through significance analysis of microarrays, 52 genes involved in important pathways related to cancer were differentially expressed in the plasma cells of healthy subjects (normal plasma-cell [NPC]; n=22) and patients with stringently defined MGUS/smoldering MM (n=24) and symptomatic MM (n=351) (P<.001). Unsupervised hierarchical clustering of 351 patients with MM, 44 with MGUS (24+20), and 16 with MM from MGUS created 2 major cluster branches, one containing 82% of the MGUS patients and the other containing 28% of the MM patients, termed MGUS-like MM (MGUS-L MM). Using the same clustering approach on an independent cohort of 214 patients with MM, 27% were found to be MGUS-L. This molecular signature, despite its association with a lower incidence of complete remission (P=.006), was associated with low-risk clinical and molecular features and superior survival (P<.01). The MGUS-L signature was also seen in plasma cells from 15 of 20 patients surviving more than 10 years after autotransplantation. These data provide insight into the molecular mechanisms of plasma-cell dyscrasias.
Figures


References
-
- Kyle RA, Therneau TM, Rajkumar SV, et al. A long-term study of prognosis in monoclonal gammopathy of undetermined significance. N Engl J Med. 2002;346:564–569. - PubMed
-
- Kyle RA, Therneau TM, Rajkumar SV, et al. Prevalence of monoclonal gammopathy of undetermined significance. N Engl J Med. 2006;354:1362–1369. - PubMed
-
- Lynch HT, Watson P, Tarantolo S, et al. Phenotypic heterogeneity in multiple myeloma families. J Clin Oncol. 2005;23:685–693. - PubMed
-
- Durie BG. The epidemiology of multiple myeloma. Semin Hematol. 2001;38:1–5. - PubMed
-
- Tilsdale JF, Stewart AK, Dickstein B, et al. Molecular and serological examination of the relationship of human herpesvirus 8 to multiple myeloma: orf 26 sequences in bone marrow stroma are not restricted to myeloma patients and other regions of the genome are not detected. Blood. 1998;92:2681–2687. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

