A second, non-canonical RNA-dependent RNA polymerase in SARS coronavirus
- PMID: 17024178
- PMCID: PMC1618104
- DOI: 10.1038/sj.emboj.7601368
A second, non-canonical RNA-dependent RNA polymerase in SARS coronavirus
Abstract
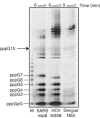
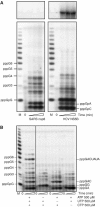
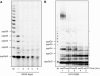
In (+) RNA coronaviruses, replication and transcription of the giant approximately 30 kb genome to produce genome- and subgenome-size RNAs of both polarities are mediated by a cognate membrane-bound enzymatic complex. Its RNA-dependent RNA polymerase (RdRp) activity appears to be supplied by non-structural protein 12 (nsp12) that includes an RdRp domain conserved in all RNA viruses. Using SARS coronavirus, we now show that coronaviruses uniquely encode a second RdRp residing in nsp8. This protein strongly prefers the internal 5'-(G/U)CC-3' trinucleotides on RNA templates to initiate the synthesis of complementary oligonucleotides of <6 residues in a reaction whose fidelity is relatively low. Distant structural homology between the C-terminal domain of nsp8 and the catalytic palm subdomain of RdRps of RNA viruses suggests a common origin of the two coronavirus RdRps, which however may have evolved different sets of catalytic residues. A parallel between the nsp8 RdRp and cellular DNA-dependent RNA primases is drawn to propose that the nsp8 RdRp produces primers utilized by the primer-dependent nsp12 RdRp.
Figures



Similar articles
-
The SARS-coronavirus nsp7+nsp8 complex is a unique multimeric RNA polymerase capable of both de novo initiation and primer extension.Nucleic Acids Res. 2012 Feb;40(4):1737-47. doi: 10.1093/nar/gkr893. Epub 2011 Oct 29. Nucleic Acids Res. 2012. PMID: 22039154 Free PMC article.
-
The RNA polymerase activity of SARS-coronavirus nsp12 is primer dependent.Nucleic Acids Res. 2010 Jan;38(1):203-14. doi: 10.1093/nar/gkp904. Epub 2009 Oct 29. Nucleic Acids Res. 2010. PMID: 19875418 Free PMC article.
-
The potential chemical structure of anti-SARS-CoV-2 RNA-dependent RNA polymerase.J Med Virol. 2020 Jun;92(6):693-697. doi: 10.1002/jmv.25761. Epub 2020 Mar 18. J Med Virol. 2020. PMID: 32167173 Free PMC article.
-
Structure-function relationships among RNA-dependent RNA polymerases.Curr Top Microbiol Immunol. 2008;320:137-56. doi: 10.1007/978-3-540-75157-1_7. Curr Top Microbiol Immunol. 2008. PMID: 18268843 Free PMC article. Review.
-
Insights into RNA synthesis, capping, and proofreading mechanisms of SARS-coronavirus.Virus Res. 2014 Dec 19;194:90-9. doi: 10.1016/j.virusres.2014.10.008. Epub 2014 Oct 17. Virus Res. 2014. PMID: 25451065 Free PMC article. Review.
Cited by
-
Biogenesis and dynamics of the coronavirus replicative structures.Viruses. 2012 Nov 21;4(11):3245-69. doi: 10.3390/v4113245. Viruses. 2012. PMID: 23202524 Free PMC article. Review.
-
A Cell-Based Reporter Assay for Screening Inhibitors of MERS Coronavirus RNA-Dependent RNA Polymerase Activity.J Clin Med. 2020 Jul 27;9(8):2399. doi: 10.3390/jcm9082399. J Clin Med. 2020. PMID: 32727069 Free PMC article.
-
Severe acute respiratory syndrome coronavirus accessory proteins 6 and 9b interact in vivo.Virus Res. 2012 Oct;169(1):282-8. doi: 10.1016/j.virusres.2012.07.012. Epub 2012 Jul 20. Virus Res. 2012. PMID: 22820404 Free PMC article.
-
A new cistron in the murine hepatitis virus replicase gene.J Virol. 2010 Oct;84(19):10148-58. doi: 10.1128/JVI.00901-10. Epub 2010 Jul 28. J Virol. 2010. PMID: 20668085 Free PMC article.
-
Murine hepatitis virus replicase protein nsp10 is a critical regulator of viral RNA synthesis.J Virol. 2007 Jun;81(12):6356-68. doi: 10.1128/JVI.02805-06. Epub 2007 Mar 28. J Virol. 2007. PMID: 17392363 Free PMC article.
References
-
- Ackermann M, Padmanabhan R (2001) De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. J Biol Chem 276: 39926–39937 - PubMed
-
- Barrette-Ng IH, Ng KK, Mark BL, Van Aken D, Cherney MM, Garen C, Kolodenko Y, Gorbalenya AE, Snijder EJ, James MN (2002) Structure of arterivirus nsp4. The smallest chymotrypsin-like proteinase with an alpha/beta C-terminal extension and alternate conformations of the oxyanion hole. J Biol Chem 277: 39960–39966 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

