Virion tails of Beet yellows virus: Coordinated assembly by three structural proteins
- PMID: 17027895
- PMCID: PMC1847569
- DOI: 10.1016/j.virol.2006.09.007
Virion tails of Beet yellows virus: Coordinated assembly by three structural proteins
Abstract
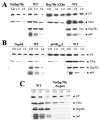
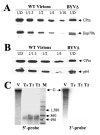
Filamentous virions of Beet yellows virus contain a long body formed by a major capsid protein and a short tail that is assembled by a minor capsid protein (CPm), an Hsp70-homolog (Hsp70h), a 64-kDa protein (p64), and a 20-kDa protein (p20). Using mutation analysis and newly developed in planta assays, here we investigate the genetic requirements for the tail assembly. We show that the inactivation of CPm dramatically reduces incorporation of both Hsp70h and p64. Furthermore, inactivation of Hsp70h prevents incorporation of p64 into virions and vice versa. Hsp70h and p64 are each required for efficient incorporation of CPm. We also show that the tails possessing normal relative amounts of CPm, Hsp70h, and p64 can be formed in the absence of the major capsid protein and p20. Similar to the tails isolated from the wild-type virions, these mutant tails encapsidate the approximately 700 nt-long, 5'-terminal segments of the viral RNA. Taken together, our results imply that CPm, Hsp70h and p64 act cooperatively to encapsidate a defined region of the closterovirus genome.
Figures


References
-
- Alzhanova DV, Hagiwara Y, Peremyslov VV, Dolja VV. Genetic analysis of the cell-to-cell movement of beet yellows closterovirus. Virology. 2000;268(1):192–200. - PubMed
-
- Atabekov JG, Rodionova NP, Karpova OV, Kozlovsky SV, Poljakov VY. The movement protein-triggered in situ conversion of potato virus X virion RNA from a nontranslatable into a translatable form. Virology. 2000;271:259–263. - PubMed
-
- Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–451. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

