Cyanobacterial ecotypes in the microbial mat community of Mushroom Spring (Yellowstone National Park, Wyoming) as species-like units linking microbial community composition, structure and function
- PMID: 17028085
- PMCID: PMC1764927
- DOI: 10.1098/rstb.2006.1919
Cyanobacterial ecotypes in the microbial mat community of Mushroom Spring (Yellowstone National Park, Wyoming) as species-like units linking microbial community composition, structure and function
Abstract
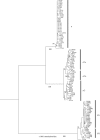
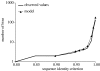
We have investigated microbial mats of alkaline siliceous hot springs in Yellowstone National Park as natural model communities to learn how microbial populations group into species-like fundamental units. Here, we bring together empirical patterns of the distribution of molecular variation in predominant mat cyanobacterial populations, theory-based modelling of how to demarcate phylogenetic clusters that correspond to ecological species and the dynamic patterns of the physical and chemical microenvironments these populations inhabit and towards which they have evolved adaptations. We show that putative ecotypes predicted by the theory-based model correspond well with distribution patterns, suggesting populations with distinct ecologies, as expected of ecological species. Further, we show that increased molecular resolution enhances our ability to detect ecotypes in this way, though yet higher molecular resolution is probably needed to detect all ecotypes in this microbial community.
Figures






References
-
- Allewalt J.A, Bateson M.M, Revsbech N.P, Slack K, Ward D.M. Temperature and light adaptations of Synechococcus isolates from the microbial mat community in Octopus Spring, Yellowstone National Park. Appl. Environ. Microbiol. 2006;72:544–550. doi:10.1128/AEM.72.1.544-550.2006 - DOI - PMC - PubMed
-
- Bryant, D. A., et al Submitted. A phototrophic acidobacterium discovered by metagenomics.
-
- Cohan F.M. What are bacterial species? Ann. Rev. Microbiol. 2002;56:457–487. doi:10.1146/annurev.micro.56.012302.160634 - DOI - PubMed
-
- Cohan F.M. Toward a conceptual and operational union of bacterial systematics, ecology, and evolution. Phil. Trans. R. Soc. B. 2006;361:1985–1996. doi:10.1098/rstb.2006.1918 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
