Expression of C-terminal deleted p53 isoforms in neuroblastoma
- PMID: 17028100
- PMCID: PMC1636465
- DOI: 10.1093/nar/gkl619
Expression of C-terminal deleted p53 isoforms in neuroblastoma
Abstract
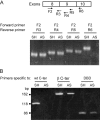
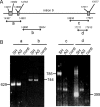
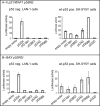
The tumor suppressor gene, p53, is rarely mutated in neuroblastomas (NB) at the time of diagnosis, but its dysfunction could result from a nonfunctional conformation or cytoplasmic sequestration of the wild-type p53 protein. However, p53 mutation, when it occurs, is found in NB tumors with drug resistance acquired over the course of chemotherapy. As yet, no study has been devoted to the function of the specific p53 mutants identified in NB cells. This study includes characterization and functional analysis of p53 expressed in eight cell lines: three wild-type cell lines and five cell lines harboring mutations. We identified two transcription-inactive p53 variants truncated in the C-terminus, one of which corresponded to the p53beta isoform recently identified in normal tissue by Bourdon et al. [J. C. Bourdon, K. Fernandes, F. Murray-Zmijewski, G. Liu, A. Diot, D. P. Xirodimas, M. K. Saville and D. P. Lane (2005) Genes Dev., 19, 2122-2137]. Our results show, for the first time, that the p53beta isoform is the only p53 species to be endogenously expressed in the human NB cell line SK-N-AS, suggesting that the C-terminus truncated p53 isoforms may play an important role in NB tumor development.
Figures






References
-
- Hainaut P., Hollstein M. p53 and human cancer: the first ten thousand mutations. Adv. Cancer Res. 2000;77:81–137. - PubMed
-
- Soussi T., Beroud C. Assessing TP53 status in human tumours to evaluate clinical outcome. Nature Rev. Cancer. 2001;1:233–240. - PubMed
-
- Beroud C., Soussi T. The UMD-p53 database: new mutations and analysis tools. Hum. Mutat. 2003;21:176–181. - PubMed
-
- Soussi T., Kato S., Levy P.P., Ishioka C. Reassessment of the TP53 mutation database in human disease by data mining with a library of TP53 missense mutations. Hum. Mutat. 2005;25:6–17. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

