A posttranslationally regulated protease, VheA, is involved in the liberation of juveniles from parental spheroids in Volvox carteri
- PMID: 17028206
- PMCID: PMC1626617
- DOI: 10.1105/tpc.106.041343
A posttranslationally regulated protease, VheA, is involved in the liberation of juveniles from parental spheroids in Volvox carteri
Abstract
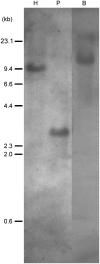
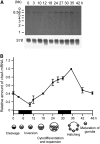
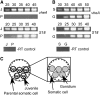
The lineage of volvocine algae includes unicellular Chlamydomonas and multicellular Volvox in addition to their colonial relatives intermediate in size and cell number. In an asexual life cycle, daughter cells of Chlamydomonas hatch from parental cell walls soon after cell division, while Volvox juveniles are released from parental spheroids after the completion of various developmental events required for the survival of multicellular juveniles. Thus, heterochronic change in the timing of hatching is considered to have played an important role in the evolution of multicellularity in volvocine algae. To study the hatching process in Volvox carteri, we purified a 125-kD Volvox hatching enzyme (VheA) from a culture medium with enzymatic activity to degrade the parental spheroids. The coding region of vheA contains a prodomain with a transmembrane segment, a subtilisin-like Ser protease domain, and a functionally unknown domain, although purified 125-kD VheA does not contain a prodomain. While 143-kD VheA with a prodomain is synthesized long before the hatching stage, 125-kD VheA is released into the culture medium during hatching due to cleavage processing at the site between the prodomain and the subtilisin-like Ser protease domain, indicating that posttranslational regulation is involved in the determination of the timing of hatching.
Figures






References
-
- Aono, N., Inoue, T., and Shiraishi, H. (2005). Genes specifically expressed in sexually differentiated female spheroids of Volvox carteri. J. Biochem. (Tokyo) 138 375–382. - PubMed
-
- Araki, K., Fujikawa, N., Nakayama, I., Nagoya, H., and Onozato, H. (1996). Early expression of a hatching enzyme gene in masu salmon (Oncorhynchus masou) embryos. Can. J. Fish. Aquat. Sci. 53 509–512.
-
- Bendtsen, J.D., Nielsen, H., von Heijne, G., and Brunak, S. (2004). Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340 783–795. - PubMed
-
- Burge, C., and Karlin, S. (1997). Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268 78–94. - PubMed
-
- Cheng, Q., Pappas, V., Hallmann, A., and Miller, S.M. (2005). Hsp70A and GlsA interact as partner chaperones to regulate asymmetric division in Volvox. Dev. Biol. 286 537–548. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

