An integrated in silico gene mapping strategy in inbred mice
- PMID: 17028314
- PMCID: PMC1774989
- DOI: 10.1534/genetics.106.065359
An integrated in silico gene mapping strategy in inbred mice
Abstract
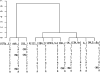
In recent years in silico analysis of common laboratory mice has been introduced and subsequently applied, in slightly different ways, as a methodology for gene mapping. Previously we have demonstrated some limitation of the methodology due to sporadic genetic correlations across the genome. Here, we revisit the three main aspects that affect in silico analysis. First, we report on the use of marker maps: we compared our existing 20,000 SNP map to the newly released 140,000 SNP map. Second, we investigated the effect of varying strain numbers on power to map QTL. Third, we introduced a novel statistical approach: a cladistic analysis, which is well suited for mouse genetics and has increased flexibility over existing in silico approaches. We have found that in our examples of complex traits, in silico analysis by itself does fail to uniquely identify quantitative trait gene (QTG)-containing regions. However, when combined with additional information, it may significantly help to prioritize candidate genes. We therefore recommend using an integrated work flow that uses other genomic information such as linkage regions, regions of shared ancestry, and gene expression information to obtain a list of candidate genes from the genome.
Figures











References
-
- Adams, D. J., E. T. Dermitzakis, T. Cox, J. Smith, R. Davies et al., 2005. Complex haplotypes, copy number polymorphisms and coding variation in two recently divergent mouse strains. Nat. Genet. 37: 532–536. - PubMed
-
- Cervino, A. C., G. Li, S. Edwards, J. Zhu, C. Laurie et al., 2005. Integrating QTL and high-density SNP analyses in mice to identify Insig2 as a susceptibility gene for plasma cholesterol levels. Genomics 86: 505–517. - PubMed
-
- Cervino, A. C., M. Gosink, M. Fallahi, B. Pascal, C. Mader et al., 2006. A comprehensive mouse IBD database for the efficient localization of quantitative trait loci. Mamm. Genome 17: 565–574. - PubMed
-
- Darvasi, A., 2001. In silico mapping of mouse quantitative trait loci. Science 294: 2423. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

